5OKC
 
 | |
6EXO
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EX8
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-~{N}-[1-(aminomethyl)cyclopropyl]-19-chloranyl-5-oxidanylidene-9-(trifluoromethyl)-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-07 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
1N2C
 
 | | NITROGENASE COMPLEX FROM AZOTOBACTER VINELANDII STABILIZED BY ADP-TETRAFLUOROALUMINATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schindelin, H, Kisker, C, Rees, D.C. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction.
Nature, 387, 1997
|
|
6EXQ
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-8-methoxy-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
1OGP
 
 | | The crystal structure of plant sulfite oxidase provides insight into sulfite oxidation in plants and animals | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(VI), CESIUM ION, GLYCEROL, ... | | Authors: | Schrader, N, Fischer, K, Theis, K, Mendel, R.R, Schwarz, G, Kisker, C. | | Deposit date: | 2003-05-08 | | Release date: | 2003-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Plant Sulfite Oxidase Provides Insights Into Sulfite Oxidation in Plants and Animals
Structure, 11, 2003
|
|
6H4J
 
 | | Usp25 catalytic domain | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4I
 
 | | Usp28 catalytic domain apo | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4H
 
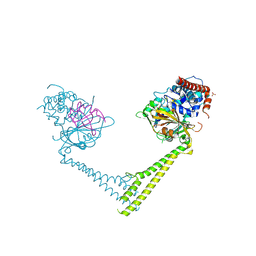 | | Usp28 catalytic domain variant E593D in complex with UbPA | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28, ... | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4K
 
 | | Structure of the Usp25 C-terminal domain | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
1Q9X
 
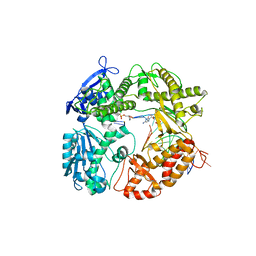 | | Crystal structure of Enterobacteria phage RB69 gp43 DNA polymerase complexed with tetrahydrofuran containing DNA | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 2',3'-DIDEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Freisinger, E, Grollman, A.P, Miller, H, Kisker, C. | | Deposit date: | 2003-08-26 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Lesion (in)tolerance reveals insights into DNA replication fidelity.
Embo J., 23, 2004
|
|
1QSG
 
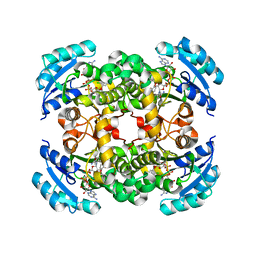 | | CRYSTAL STRUCTURE OF ENOYL REDUCTASE INHIBITION BY TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN, ... | | Authors: | Stewart, M.J, Parikh, S, Xiao, G, Tonge, P.J, Kisker, C. | | Deposit date: | 1999-06-21 | | Release date: | 1999-07-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis and mechanism of enoyl reductase inhibition by triclosan.
J.Mol.Biol., 290, 1999
|
|
1MJ3
 
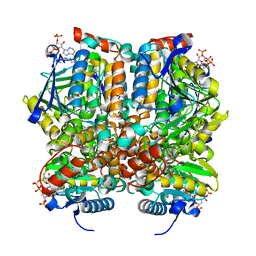 | | Crystal Structure Analysis of rat enoyl-CoA hydratase in complex with hexadienoyl-CoA | | Descriptor: | ENOYL-COA HYDRATASE, MITOCHONDRIAL, HEXANOYL-COENZYME A | | Authors: | Bell, A.F, Feng, Y, Hofstein, H.A, Parikh, S, Wu, J, Rudolph, M.J, Kisker, C, Tonge, P.J. | | Deposit date: | 2002-08-26 | | Release date: | 2002-09-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stereoselectivity of Enoyl-CoA Hydratase Results from Preferential Activation of
One of Two Bound Substrate Conformers
Chem.Biol., 9, 2002
|
|
2TRT
 
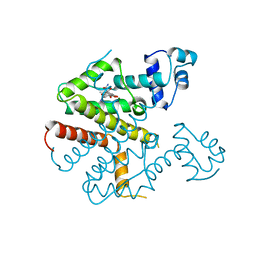 | | TETRACYCLINE REPRESSOR CLASS D | | Descriptor: | MAGNESIUM ION, TETRACYCLINE, TETRACYCLINE REPRESSOR CLASS D | | Authors: | Hinrichs, W, Kisker, C, Saenger, W. | | Deposit date: | 1994-03-04 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance.
Science, 264, 1994
|
|
2WGD
 
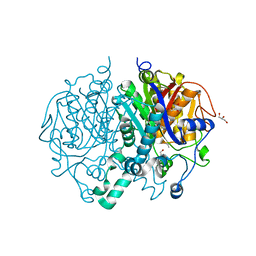 | | Crystal structure of KasA of Mycobacterium tuberculosis | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Luckner, S.R, Kisker, C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structures of Mycobacterium Tuberculosis Kasa Show Mode of Action within Cell Wall Biosynthesis and its Inhibition by Thiolactomycin
Structure, 17, 2009
|
|
2WGG
 
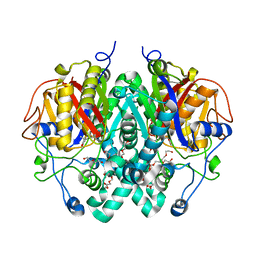 | |
2WGF
 
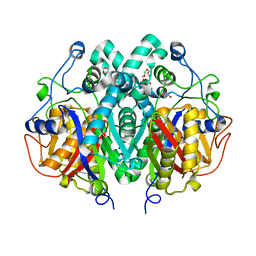 | | Crystal structure of Mycobacterium tuberculosis C171Q KasA variant | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Luckner, S.R, Kisker, C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Mycobacterium Tuberculosis Kasa Show Mode of Action within Cell Wall Biosynthesis and its Inhibition by Thiolactomycin
Structure, 17, 2009
|
|
2W3R
 
 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in complex with hypoxanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2W55
 
 | | Crystal Structure of Xanthine Dehydrogenase (E232Q variant) from Rhodobacter capsulatus in Complex with Hypoxanthine | | Descriptor: | BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2W54
 
 | | Crystal Structure of Xanthine Dehydrogenase from Rhodobacter capsulatus in Complex with Bound Inhibitor Pterin-6-aldehyde | | Descriptor: | 6-HYDROXYMETHYLPTERIN, BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2VSF
 
 | | Structure of XPD from Thermoplasma acidophilum | | Descriptor: | CALCIUM ION, DNA REPAIR HELICASE RAD3 RELATED PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Kuper, J, Wolski, S.C, Truglio, J.J, Kisker, C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Fes Cluster-Containing Nucleotide Excision Repair Helicase Xpd.
Plos Biol., 6, 2008
|
|
2W3S
 
 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in Complex with Xanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2WGE
 
 | |
2X23
 
 | | crystal structure of M. tuberculosis InhA inhibited by PT70 | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, DIMETHYL SULFOXIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], ... | | Authors: | Luckner, S.R, Liu, N, am Ende, C.W, Tonge, P.J, Kisker, C. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | A Slow, Tight Binding Inhibitor of Inha, the Enoyl-Acyl Carrier Protein Reductase from Mycobacterium Tuberculosis.
J.Biol.Chem., 285, 2010
|
|
4BKR
 
 | | Enoyl-ACP reductase from Yersinia pestis (wildtype, removed Histag) with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
