7MJL
 
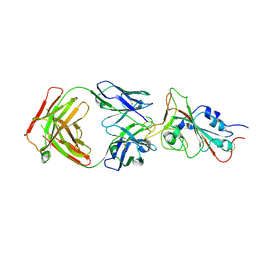 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (focused refinement of RBD and Fab ab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, Fab ab1 Light Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJN
 
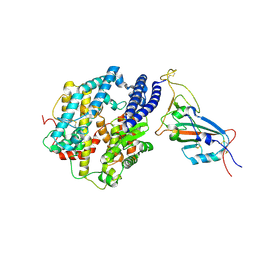 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJK
 
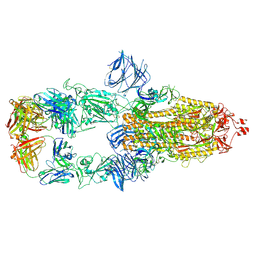 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
5NO3
 
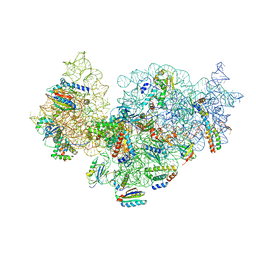 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate without uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
8GS8
 
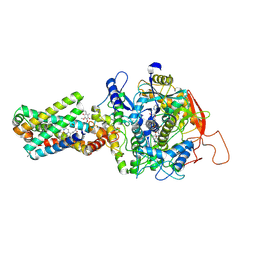 | | cryo-EM structure of the human respiratory complex II | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Du, Z, Zhou, X, Lai, Y, Xu, J, Zhang, Y, Zhou, S, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-09-05 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the human respiratory complex II.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5NO4
 
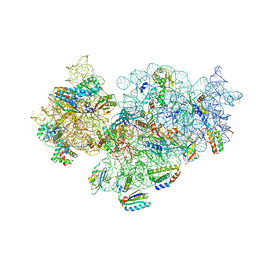 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate with uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5NO2
 
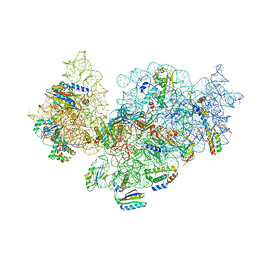 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
6VFO
 
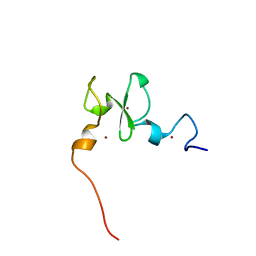 | | Solution structure of the PHD of mouse UHRF1 (NP95) | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Lemak, A, Houliston, S, Duan, S, Arrowsmith, C.H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
6VEE
 
 | |
6VED
 
 | | Solution structure of the TTD and linker region of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1 | | Authors: | Lemak, A, Houliston, S, Duan, S, Ong, M.S, Arrowsmith, C.H. | | Deposit date: | 2019-12-31 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
5W7G
 
 | | An envelope of a filamentous hyperthermophilic virus carries lipids in a horseshoe conformation | | Descriptor: | DNA (253-MER), ORF132, ORF140 | | Authors: | Kasson, P, DiMaio, F, Yu, X, Lucas-Staat, S, Krupovic, M, Schouten, S, Prangishvili, D, Egelman, E. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Model for a novel membrane envelope in a filamentous hyperthermophilic virus.
Elife, 6, 2017
|
|
3OWG
 
 | | Crystal structure of vaccinia virus Polyadenylate polymerase(vp55) | | Descriptor: | Poly(A) polymerase catalytic subunit | | Authors: | Li, C, Li, H, Zhou, S, Gershon, P.D, Poulos, T.L. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Domain-level rocking motion within a polymerase that translocates on single-stranded nucleic acid.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7LQT
 
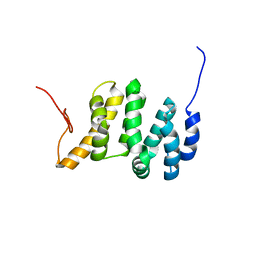 | | Solution NMR structure of the PNUTS amino-terminal Domain fused to Myc Homology Box 0 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10,Myc proto-oncogene protein fusion | | Authors: | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
8J4I
 
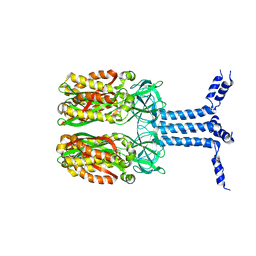 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
8ODQ
 
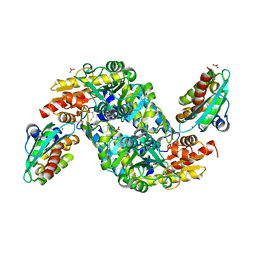 | | SufS-SufU complex from Mycobacterium tuberculosis | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Cysteine desulfurase, NITRATE ION, ... | | Authors: | Elchennawi, I, Carpentier, P, Caux, C, Ponge, M, Ollagnier de Choudens, S. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium tuberculosis Zinc SufU-SufS Complex.
Biomolecules, 13, 2023
|
|
3ROF
 
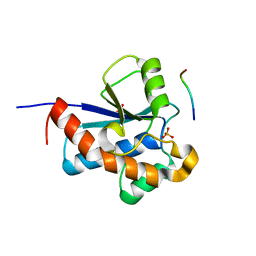 | | Crystal Structure of the S. aureus Protein Tyrosine Phosphatase PtpA | | Descriptor: | Expression tag cleaved from protein-tyrosine-phosphatase ptpA, Low molecular weight protein-tyrosine-phosphatase ptpA, PHOSPHATE ION | | Authors: | Grundner, C, Chou, S, Engel, K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure and substrate recognition of the Staphylococcus aureus protein tyrosine phosphatase PtpA.
J.Mol.Biol., 413, 2011
|
|
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | Descriptor: | HEEH_rd4_0097 | | Authors: | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
1J4Q
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T192) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
2VH6
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with biaryl P4 motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-{(3S)-1-[3-fluoro-2'-(methylsulfonyl)biphenyl-4-yl]-2-oxopyrrolidin-3-yl}ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Chung, C.W, Convery, M.A, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A.N, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E. | | Deposit date: | 2007-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Biaryl P4 Motifs.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1K3Q
 
 | | NMR structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T192) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3N
 
 | | NMR Structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T155) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
4KT3
 
 | | Structure of a type VI secretion system effector-immunity complex from Pseudomonas protegens | | Descriptor: | Putative lipoprotein, Uncharacterized protein | | Authors: | Whitney, J.C, Chou, S, Gardiner, T.E, Mougous, J.D. | | Deposit date: | 2013-05-19 | | Release date: | 2013-07-31 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (1.4362 Å) | | Cite: | Identification, Structure, and Function of a Novel Type VI Secretion Peptidoglycan Glycoside Hydrolase Effector-Immunity Pair.
J.Biol.Chem., 288, 2013
|
|
