7DHV
 
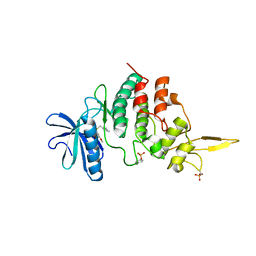 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 8 | | Descriptor: | 2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine-4-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-17 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DJO
 
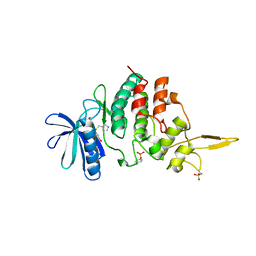 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 17 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-[[(3S)-pyrrolidin-3-yl]methylsulfanyl]acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DL6
 
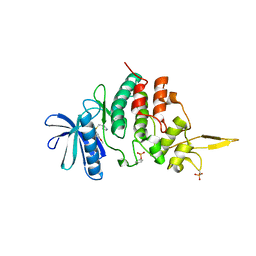 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 18 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-[[(3R)-pyrrolidin-3-yl]methylsulfanyl]acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DXI
 
 | |
7E5E
 
 | | Crystal structure of GDP-bound GNAS in complex with the cyclic peptide inhibitor GD20 | | Descriptor: | CHLORIDE ION, GD20, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hu, Q, Dai, S, Shokat, K.M. | | Deposit date: | 2021-02-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | State-selective modulation of heterotrimeric G alpha s signaling with macrocyclic peptides.
Cell, 185, 2022
|
|
7DVM
 
 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
7CYN
 
 | | Cryo-EM structure of human TLR7 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DTZ
 
 | | FGFR4 complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[5-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]methoxy]pyrimidin-2-yl]amino]-3-methyl-phenyl]-2-fluoranyl-prop-2-enamide, SULFATE ION | | Authors: | Chen, X.J, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-01-07 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Investigation of Covalent Warheads in the Design of 2-Aminopyrimidine-based FGFR4 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7EGV
 
 | | Acetolactate Synthase from Trichoderma harzianum with inhibitor harzianic acid | | Descriptor: | (2S)-3-methyl-2-[[(2S,4R)-1-methyl-4-[(2E,4E)-octa-2,4-dienoyl]-3,5-bis(oxidanylidene)pyrrolidin-2-yl]methyl]-2-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Zang, X, Xie, L, Chen, M, Tang, Y, Zhou, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Harzianic Acid from Trichoderma afroharzianum Is a Natural Product Inhibitor of Acetohydroxyacid Synthase.
J.Am.Chem.Soc., 2021
|
|
7EHE
 
 | | Acetolactate Synthase from Trichoderma harzianum | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zang, X, Tang, Y, Zhou, J. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Harzianic Acid from Trichoderma afroharzianum Is a Natural Product Inhibitor of Acetohydroxyacid Synthase.
J.Am.Chem.Soc., 2021
|
|
7EYA
 
 | |
7EY0
 
 | |
7EZV
 
 | |
7EY4
 
 | | Local CryoEM of the SARS-CoV-2 S6PV2 in complex with BD-667 | | Descriptor: | BD-667 H, BD-667 L, Spike glycoprotein, ... | | Authors: | Liu, P.L. | | Deposit date: | 2021-05-29 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants.
Cell Res., 31, 2021
|
|
7E4Z
 
 | | Crystal structure of tubulin in complex with Maytansinol | | Descriptor: | (1R,2S,3S,5S,6S,16E,18E,20R,21S)-11-chloro-6,21-dihydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4Q
 
 | | Crystal structure of tubulin in complex with L-DM1-SMe | | Descriptor: | (1S,2R,3R,5R,6R,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-(methyldisulfanyl)propanoyl]amino}propanoate (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4R
 
 | | Crystal structure of tubulin in complex with D-DM1-SMe | | Descriptor: | (10E,12E)-86-chloro-14-hydroxy-85,14-dimethoxy-33,2,7,10-tetramethyl-12,6-dioxo-7-aza-1(6,4)-oxazinana-3(2,3)-oxirana-8(1,3)-benzenacyclotetradecaphane-10,12-dien-4-yl N-methyl-N-(3-(methylsulfinothioyl)propanoyl)-D-alaninate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7C77
 
 | | Cryo-EM structure of mouse TLR3 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-05-23 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7C76
 
 | | Cryo-EM structure of human TLR3 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-05-23 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DPX
 
 | |
7FTP
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 6-(4-chlorophenyl)-10,11-dimethoxy-7-methyl-2,4,6-triazatricyclo[7.3.1.05,13]trideca-1(12),2,4,7,9(13),10-hexaene | | Descriptor: | 4-(4-chlorophenyl)-7,8-dimethoxy-5-methyl-4H-pyrido[2,3,4-de]quinazoline, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Zhang, Z, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
