1DO9
 
 | | SOLUTION STRUCTURE OF OXIDIZED MICROSOMAL RABBIT CYTOCHROME B5. FACTORS DETERMINING THE HETEROGENEOUS BINDING OF THE HEME. | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Rosato, A, Scacchieri, S. | | Deposit date: | 1999-12-20 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized microsomal rabbit cytochrome b5. Factors determining the heterogeneous binding of the heme.
Eur.J.Biochem., 267, 2000
|
|
6FY3
 
 | | Crystal structure of a V2-directed, RV144 vaccine-like antibody from HIV-1 infection, CAP228-3D, bound to a heterologous V2 peptide | | Descriptor: | CAP228-3D Heavy Chain, CAP228-3D Light Chain, CAP45 V2 peptide | | Authors: | Wibmer, C.K, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Common helical V1V2 conformations of HIV-1 Envelope expose the alpha 4 beta 7 binding site on intact virions.
Nat Commun, 9, 2018
|
|
3TS5
 
 | | Crystal Structure of a Light Chain Domain of Scallop Smooth Muscle Myosin | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Kumar, V.S.S, O'Neall-Hennessey, E, Reshetnikova, L, Brown, J.H, Robinson, H, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2011-09-12 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal structure of a phosphorylated light chain domain of scallop smooth-muscle Myosin.
Biophys.J., 101, 2011
|
|
4D47
 
 | | X-ray structure of the levansucrase from Erwinia amylovora | | Descriptor: | LEVANSUCRASE, alpha-D-glucopyranose, beta-D-fructofuranose | | Authors: | Wuerges, J, Caputi, L, Cianci, M, Benini, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | The Crystal Structure of Erwinia Amylovora Levansucrase Provides a Snapshot of the Products of Sucrose Hydrolysis Trapped Into the Active Site.
J.Struct.Biol., 191, 2015
|
|
8B9P
 
 | | ACE2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-GLY-ARG-GLN-PHE-CYS-HIS-THR-LEU-MET-PRO-ARG-HIS-LEU-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
5O9G
 
 | | Structure of nucleosome-Chd1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Farnung, L, Vos, S.M, Wigge, C, Cramer, P. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Nucleosome-Chd1 structure and implications for chromatin remodelling.
Nature, 550, 2017
|
|
8BJI
 
 | | chimera of ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo fused to a proline-Rich-Domain (PRD) and profilin, bound to ADP-Mg-actin and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BR1
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
5O15
 
 | | Crystal structure of bifunctional dehydratase-cyclase domain in ambruticin biosynthesis | | Descriptor: | AmbC, GLYCEROL | | Authors: | Sung, K.H, Berkhan, G, Hollmann, T, Wagner, L, Hahn, F, Blankenfeldt, W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.174 Å) | | Cite: | Insights into the Dual Activity of a Bifunctional Dehydratase-Cyclase Domain.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8BO1
 
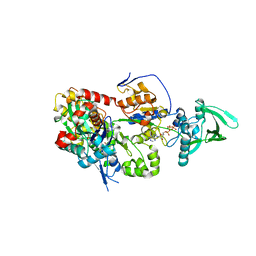 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AZIDE ION, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
5O1M
 
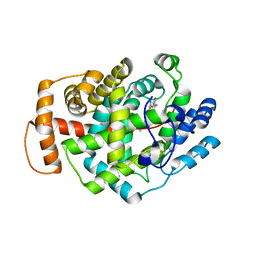 | | Structure of Latex Clearing Protein LCP in the closed state | | Descriptor: | 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Rubber oxygenase | | Authors: | Ilcu, L, Roether, W, Birke, J, Brausemann, A, Einsle, O, Jendrossek, D. | | Deposit date: | 2017-05-18 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of Latex Clearing Protein (Lcp) Provides Insight into the Enzymatic Cleavage of Rubber.
Sci Rep, 7, 2017
|
|
8SLP
 
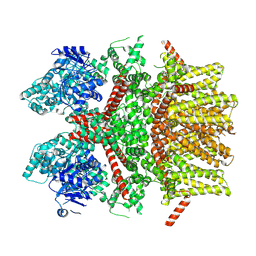 | | Cryo-EM structure of the rat TRPM5 channel in 2mM calcium, high-2 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UDU
 
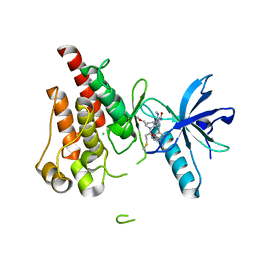 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8CLL
 
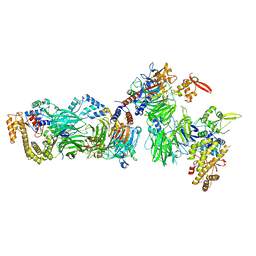 | | Structural insights into human TFIIIC promoter recognition | | Descriptor: | General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 2, General transcription factor 3C polypeptide 4, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
1PJO
 
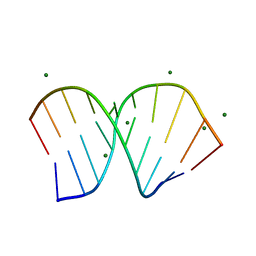 | | Crystal Structure of an RNA/DNA hybrid of HIV-1 PPT | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', MAGNESIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An Unusual Sugar Conformation in the Structure of an RNA/DNA
Decamer of the Polypurine Tract May Affect Recognition by RNase H
J.Mol.Biol., 334, 2003
|
|
8UW4
 
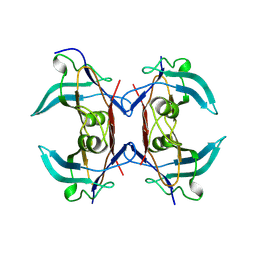 | |
5ZEC
 
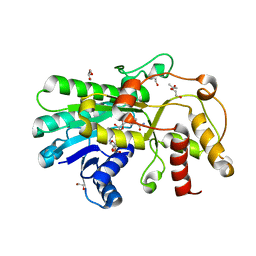 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Q136N/F161V/S196G/E214G/S237C) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wang, Y, ZHou, J.Y, Hou, X.D, Xu, G.C, Rao, Y.J, Wu, L, Zhou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
1FZD
 
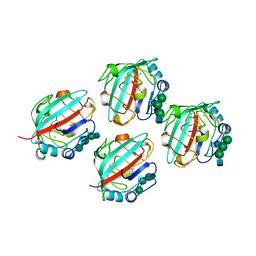 | | STRUCTURE OF RECOMBINANT ALPHAEC DOMAIN FROM HUMAN FIBRINOGEN-420 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Spraggon, G, Applegate, D, Everse, S.J, Zhang, J.-Z, Veerapandian, L, Redman, C, Doolittle, R.F, Grieninger, G. | | Deposit date: | 1998-06-22 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a recombinant alphaEC domain from human fibrinogen-420.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6EWT
 
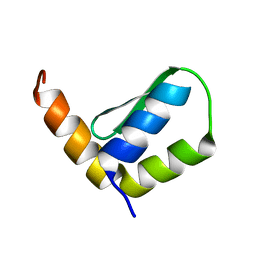 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | Descriptor: | NRPS Kj12B-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
5O4J
 
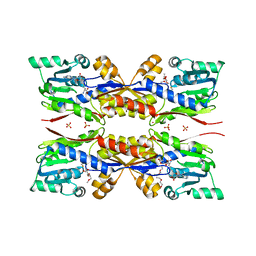 | | HcgC from Methanococcus maripaludis cocrystallized with SAH and pyridinol | | Descriptor: | (3~{E})-3-[(~{E})-3-oxidanylprop-2-enoyl]iminopropanoic acid, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8UDT
 
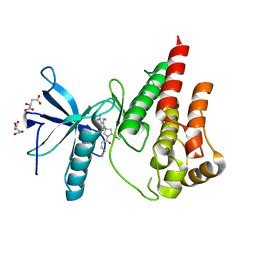 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
3TZW
 
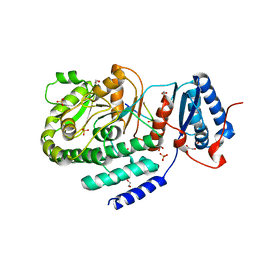 | | Crystal structure of a fragment containing the acyltransferase domain of Pks13 from Mycobacterium tuberculosis in the orthorhombic apoform at 2.6 A | | Descriptor: | 1,2-ETHANEDIOL, 12-mer peptide, Polyketide synthase PKS13, ... | | Authors: | Bergeret, F, Pedelacq, J.D, Mourey, L, Bon, C. | | Deposit date: | 2011-09-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase pks13
J.Biol.Chem., 287, 2012
|
|
6F3X
 
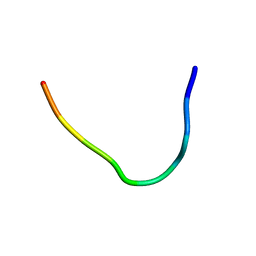 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide in frozen DDM/CHS detergent micelle solution determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
5O5U
 
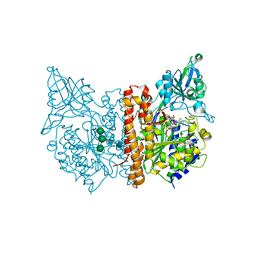 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[[(2~{S})-2-[[4-[[[(2~{S})-2-[[(2~{R})-2-[(6-fluoranylpyridin-3-yl)carbonylamino]-3-sulfo-propanoyl]amino]-3-sulfo-propanoyl]amino]methyl]phenyl]carbonylamino]-3-naphthalen-2-yl-propanoyl]amino]-1-oxidanyl-1-oxidanylidene-hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Novakova, Z, Barinka, C, Motlova, L. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027
To Be Published
|
|
6F3Y
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide bound to human Bradykinin 1 Receptor (B1R) determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
