4PQA
 
 | | Crystal Structure of succinyl-diaminopimelate desuccinylase from Neisseria meningitidis MC58 in complex with the Inhibitor Captopril | | Descriptor: | L-CAPTOPRIL, SULFATE ION, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Nocek, B, Starus, A, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
2FE7
 
 | | The crystal structure of a probable N-acetyltransferase from Pseudomonas aeruginosa | | Descriptor: | probable N-acetyltransferase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a N-acetyltransferase from Pseudomonas aeruginosa
To be Published
|
|
2FIW
 
 | | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris | | Descriptor: | ACETYL COENZYME *A, GCN5-related N-acetyltransferase:Aminotransferase, class-II, ... | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris
To be Published, 2006
|
|
2GTS
 
 | | Structure of Protein of Unknown Function HP0062 from Helicobacter pylori | | Descriptor: | hypothetical protein HP0062 | | Authors: | Binkowski, T.A, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hypothetical protein HP0062 from Helicobacter pylori
To be Published
|
|
4Q63
 
 | | Crystal Structure of Legionella Uncharacterized Protein Lpg0364 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-21 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Crystal Structure of Legionella Uncharacterized Protein Lpg0364
To be Published
|
|
2OEB
 
 | | The crystal structure of gene product Af1862 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein | | Authors: | Zhang, R, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The crystal structure of gene product Af1862 from Archaeoglobus fulgidus
To be Published
|
|
2FA8
 
 | | Crystal Structure of the Putative Selenoprotein W-related family Protein from Agrobacterium tumefaciens | | Descriptor: | hypothetical protein Atu0228 | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-07 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Selenoprotein W-related family Protein from Agrobacterium tumefaciens
To be Published, 2006
|
|
3DTZ
 
 | | Crystal structure of Putative Chlorite dismutase TA0507 | | Descriptor: | FORMIC ACID, Putative Chlorite dismutase TA0507 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of Putative Chlorite dismutase TA0507
To be Published
|
|
2ESN
 
 | | The crystal structure of probable transcriptional regulator PA0477 from Pseudomonas aeruginosa | | Descriptor: | probable transcriptional regulator | | Authors: | Lunin, V.V, Chang, C, Skarina, T, Gorodischenskaya, E, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of putative transcriptional regulator Pa0477 from Pseudomonas aeruginosa
To be Published
|
|
2PPX
 
 | | Crystal structure of a HTH XRE-family like protein from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein Atu1735 | | Authors: | Cuff, M.E, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a HTH XRE-family like protein from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
3DNH
 
 | | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58 | | Descriptor: | uncharacterized protein Atu2129 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58
To be Published
|
|
4E5S
 
 | |
2PHN
 
 | | Crystal structure of an amide bond forming F420-gamma glutamyl ligase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, F420-0:gamma-glutamyl ligase, ... | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an Amide Bond Forming F(420):gammagamma-glutamyl Ligase from Archaeoglobus Fulgidus - A Member of a New Family of Non-ribosomal Peptide Synthases.
J.Mol.Biol., 372, 2007
|
|
3NKD
 
 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | CRISPR-associated protein Cas1 | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
2GUH
 
 | | Crystal Structure of the Putative TetR-family Transcriptional Regulator from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Putative TetR-family transcriptional regulator | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Zheng, H, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-30 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the Putative TetR-family Transcriptional Regulator from Rhodococcus sp. RHA1
To be Published
|
|
2H0U
 
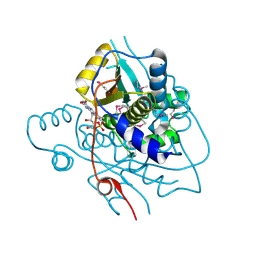 | | Crystal structure of NAD(P)H-flavin oxidoreductase from Helicobacter pylori | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-flavin oxidoreductase | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-15 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of NAD(P)H-flavin oxidoreductase from Helicobacter pylori
To be Published
|
|
2R8W
 
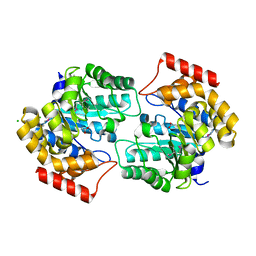 | | The crystal structure of dihydrodipicolinate synthase (Atu0899) from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETATE ION, AGR_C_1641p, CHLORIDE ION | | Authors: | Tan, K, Dong, A, Xu, X, Gu, J, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of dihydrodipicolinate synthase (Atu0899) from Agrobacterium tumefaciens str. C58.
To be Published
|
|
4QPE
 
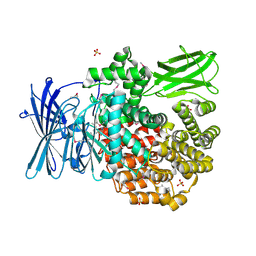 | | Crystal structure of Aminopeptidase N in complex with N-cyclohexyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION, ... | | Authors: | Nocek, B, Mulligan, R, Berlicki, L, Vassilious, S, Mucha, A, Joachimiak, A. | | Deposit date: | 2014-06-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4QME
 
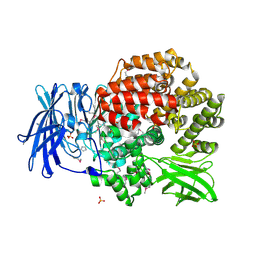 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-hPheP[CH2]Phe | | Descriptor: | (2S)-3-[(S)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Vassilious, S, Mulligan, R, Berlicki, L, Mucha, A, Joachimiak, A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4HTQ
 
 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 6.70 x 10e+11 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.Z, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4QN8
 
 | | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | BETA-MERCAPTOETHANOL, VipE | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
4QIR
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-2-(pyridin-3-yl)ethylGlyP[CH2]Phe | | Descriptor: | 3-{[(R)-1-amino-3-(pyridin-3-yl)propyl](hydroxy)phosphoryl}-(S)-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Joachimiak, A, Berlicki, L, Vassiliou, S, Mucha, A. | | Deposit date: | 2014-06-01 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
2H8O
 
 | | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens | | Descriptor: | Geranyltranstransferase | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens
To be Published
|
|
3EFE
 
 | | The crystal structure of the thiJ/pfpI family protein from Bacillus anthracis | | Descriptor: | SULFATE ION, ThiJ/pfpI family protein | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the thiJ/pfpI family protein from Bacillus anthracis
To be Published
|
|
3E9Q
 
 | | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-23 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri
To be Published
|
|
