5YEI
 
 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
5YLO
 
 | | Structural of Pseudomonas aeruginosa PA4980 | | Descriptor: | GLYCEROL, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Li, T, Peng, C.T, Li, C.C, Xiao, Q.J, He, L.H, Wang, N.Y, Bao, R. | | Deposit date: | 2017-10-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural characterization of a Delta3, Delta2-enoyl-CoA isomerase from Pseudomonas aeruginosa: implications for its involvement in unsaturated fatty acid metabolism.
J.Biomol.Struct.Dyn., 37, 2019
|
|
1D3U
 
 | | TATA-BINDING PROTEIN/TRANSCRIPTION FACTOR (II)B/BRE+TATA-BOX COMPLEX FROM PYROCOCCUS WOESEI | | Descriptor: | DNA 23-MER: BRE+TATA-BOX, DNA 24-MER: BRE+TATA-BOX, TATA-BINDING PROTEIN, ... | | Authors: | Littlefield, O, Korkhin, Y, Sigler, P.B. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the oriented assembly of a TBP/TFB/promoter complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7WQV
 
 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
8I4S
 
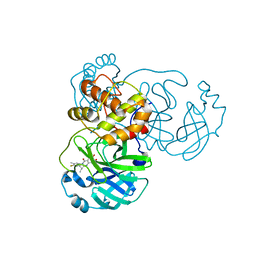 | | the complex structure of SARS-CoV-2 Mpro with D8 | | Descriptor: | 3-(4-fluoranyl-3-methyl-phenyl)-2-(2-methylpropyl)-5,6,7-tris(oxidanyl)quinazolin-4-one, ORF1a polyprotein | | Authors: | Lu, M. | | Deposit date: | 2023-01-21 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of quinazolin-4-one-based non-covalent inhibitors targeting the severe acute respiratory syndrome coronavirus 2 main protease (SARS-CoV-2 M pro ).
Eur.J.Med.Chem., 257, 2023
|
|
8JJM
 
 | |
7DK6
 
 | |
7DCC
 
 | |
7DDN
 
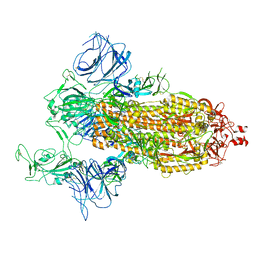 | | SARS-Cov2 S protein at open state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-29 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
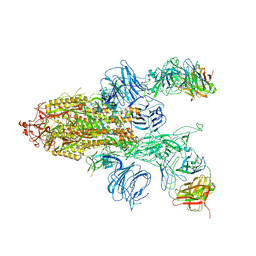
7DD2
 
 | |
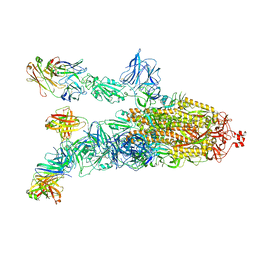
7DK4
 
 | |
7DDD
 
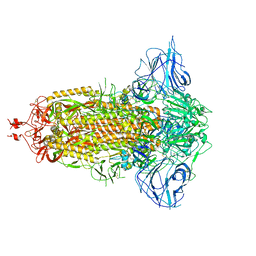 | | SARS-Cov2 S protein at close state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-28 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DCX
 
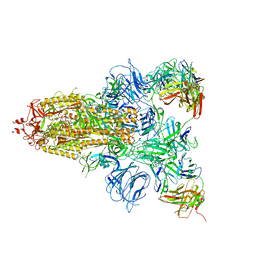 | |
7DK7
 
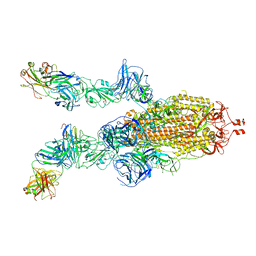 | |
7DD8
 
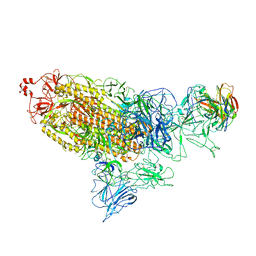 | |
7DK5
 
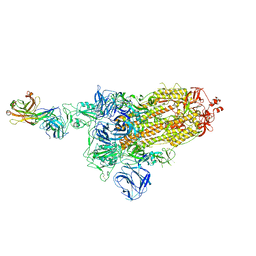 | |
7CO5
 
 | | HtrA-type protease AlgW with decapeptide | | Descriptor: | AlgW protein, IMIDAZOLE, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO3
 
 | | HtrA-type protease AlgWS227A with tripeptide | | Descriptor: | AlgW protein, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO2
 
 | | HtrA-type protease AlgW with tripeptide | | Descriptor: | AlgW protein, IMIDAZOLE, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO7
 
 | | HtrA-type protease AlgWS227A with decapeptide | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, AlgW protein, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
3LXG
 
 | | Crystal structure of rat phosphodiesterase 10A in complex with ligand WEB-3 | | Descriptor: | 2-methoxy-6,7-dimethyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Mosbacher, T, Jestel, A, Steinbacher, S. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,5-a]pyrido[3,2-e]pyrazines as a new class of phosphodiesterase 10A inhibitiors.
J.Med.Chem., 53, 2010
|
|
4FCB
 
 | | Potent and Selective Phosphodiesterase 10A Inhibitors | | Descriptor: | 3,4-dimethyl-1-propyl-7-(quinolin-2-ylmethoxy)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazines as potent and selective phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4FCD
 
 | | Potent and Selective Phosphodiesterase 10A Inhibitors | | Descriptor: | 1-(2-chlorophenyl)-6,8-dimethoxy-3-methylimidazo[5,1-c][1,2,4]benzotriazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Novel triazines as potent and selective phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3SN7
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 8-fluoro-6-methoxy-3,4-dimethyl-1-(3-methylpyridin-4-yl)imidazo[1,5-a]quinoxaline, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2011-06-28 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
