2D32
 
 | | Crystal Structure of Michaelis Complex of gamma-Glutamylcysteine Synthetase | | Descriptor: | CYSTEINE, GLUTAMIC ACID, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of efficient coupling peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
2D33
 
 | | Crystal Structure of gamma-Glutamylcysteine Synthetase Complexed with Aluminum Fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CYSTEINE, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of efficient coupling between peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
3VX8
 
 | |
3VIQ
 
 | | Crystal structure of Swi5-Sfr1 complex from fission yeast | | Descriptor: | GLYCEROL, Mating-type switching protein swi5, NITRATE ION, ... | | Authors: | Kuwabara, N, Murayama, Y, Hashimoto, H, Kokabu, Y, Ikeguchi, M, Sato, M, Mayanagi, K, Tsutsui, Y, Iwasaki, H, Shimizu, T. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Structure, 20, 2012
|
|
3VX7
 
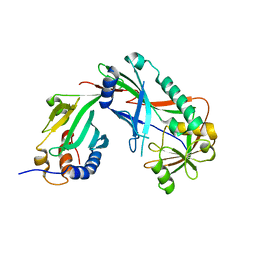 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VX6
 
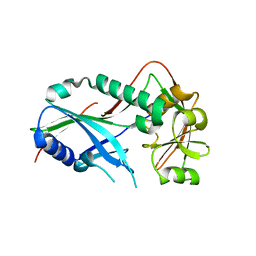 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
1IT1
 
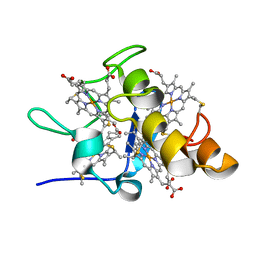 | | Solution structures of ferrocytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | HEME C, cytochrome c3 | | Authors: | Harada, E, Fukuoka, Y, Ohmura, T, Fukunishi, A, Kawai, G, Fujiwara, T, Akutsu, H. | | Deposit date: | 2001-12-29 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Redox-coupled conformational alternations in cytochrome c(3) from D. vulgaris Miyazaki F on the basis of its reduced solution structure.
J.Mol.Biol., 319, 2002
|
|
5Z9W
 
 | | Ebola virus nucleoprotein-RNA complex | | Descriptor: | Ebolavirus nucleoprotein (residues 19-406), RNA (6-MER) | | Authors: | Sugita, Y, Matsunami, H, Kawaoka, Y, Noda, T, Wolf, M. | | Deposit date: | 2018-02-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 angstrom resolution.
Nature, 563, 2018
|
|
1X03
 
 | | Crystal structure of endophilin BAR domain | | Descriptor: | SH3-containing GRB2-like protein 2 | | Authors: | Masuda, M, Takeda, S, Sone, M, Kamioka, Y, Mori, H, Mochizuki, N. | | Deposit date: | 2005-03-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms
Embo J., 25, 2006
|
|
1X04
 
 | | Crystal structure of endophilin BAR domain (mutant) | | Descriptor: | SH3-containing GRB2-like protein 2 | | Authors: | Masuda, M, Takeda, S, Sone, M, Kamioka, Y, Mori, H, Mochizuki, N. | | Deposit date: | 2005-03-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms
Embo J., 25, 2006
|
|
3VMY
 
 | |
3VP7
 
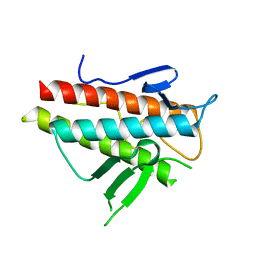 | | Crystal structure of the beta-alpha repeated, autophagy-specific (BARA) domain of Vps30/Atg6 | | Descriptor: | Vacuolar protein sorting-associated protein 30 | | Authors: | Noda, N.N, Kobayashi, T, Adachi, W, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy.
J.Biol.Chem., 287, 2012
|
|
3W1S
 
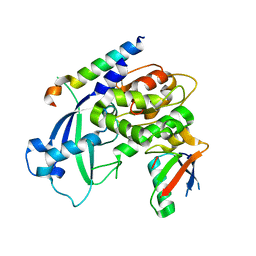 | | Crystal structure of Saccharomyces cerevisiae Atg12-Atg5 conjugate bound to the N-terminal domain of Atg16 | | Descriptor: | Autophagy protein 16, Autophagy protein 5, Ubiquitin-like protein ATG12 | | Authors: | Noda, N.N, Fujioka, Y, Hanada, T, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation
Embo Rep., 14, 2013
|
|
3VN0
 
 | |
3VMZ
 
 | |
3WH2
 
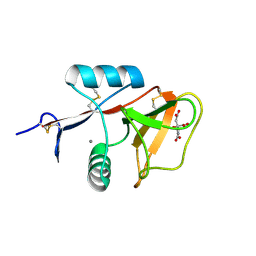 | | Human Mincle in complex with citrate | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION, CITRATE ANION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WHD
 
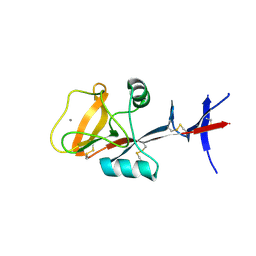 | | C-type lectin, human MCL | | Descriptor: | C-type lectin domain family 4 member D, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WH3
 
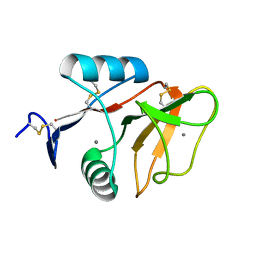 | | human Mincle, ligand free form | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7D36
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(3S)-1-amino-5-fluoro-3-methyl-3,4-dihydro-2,6-naphthyridin-3-yl]-4-fluorophenyl}-5-cyano-3-methylpyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Nakahara, K, Mitsuoka, Y, Kasuya, S, Yamamoto, T, Yamamoto, S, Ito, H, Kido, Y, Kusakabe, K.I. | | Deposit date: | 2020-09-18 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Balancing potency and basicity by incorporating fluoropyridine moieties: Discovery of a 1-amino-3,4-dihydro-2,6-naphthyridine BACE1 inhibitor that affords robust and sustained central A beta reduction.
Eur.J.Med.Chem., 216, 2021
|
|
7CCO
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lanthanum ion (La3+) | | Descriptor: | LANTHANUM (III) ION, LBT3 | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
7CCN
 
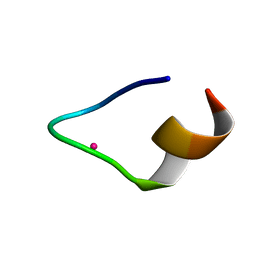 | | The binding structure of a lanthanide binding tag (LBT3) with lutetium ion (Lu3+) | | Descriptor: | LBT3, LUTETIUM (III) ION | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
1PD2
 
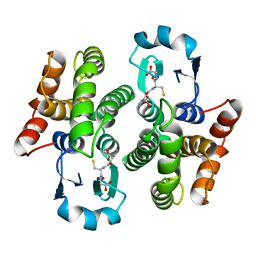 | |
