6K3F
 
 | |
7OXF
 
 | |
2LZO
 
 | | Spatial structure of Pi-AnmTX Ugr 9a-1 | | Descriptor: | UGTX | | Authors: | Mineev, K, Arseniev, A. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sea Anemone Peptide with Uncommon beta-Hairpin Structure Inhibits Acid-sensing Ion Channel 3 (ASIC3) and Reveals Analgesic Activity.
J.Biol.Chem., 288, 2013
|
|
2MKA
 
 | |
2N5S
 
 | |
2VV1
 
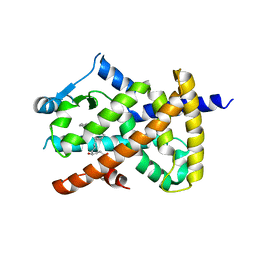 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VSR
 
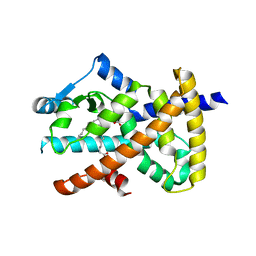 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
4URS
 
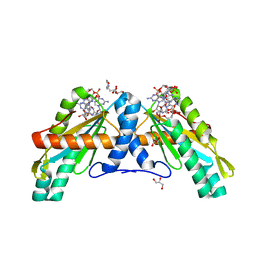 | | Crystal Structure of GGDEF domain from T.maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE, ... | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
7AY8
 
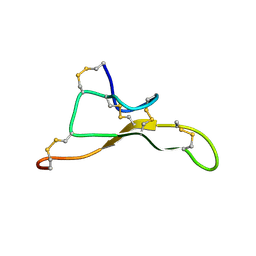 | | NMR solution structure of Tbo-IT2 | | Descriptor: | Tbo-IT2 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Korolkova, Y.A, Maleeva, E.E, Andreev, Y.A, Kozlov, S.A, Arseniev, A.S. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold.
Toxins, 13, 2021
|
|
7AV6
 
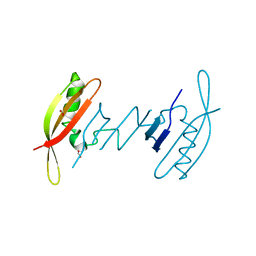 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
7E69
 
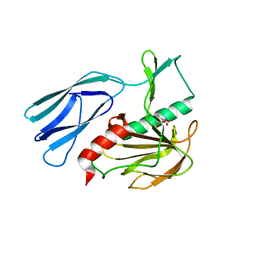 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-3 | | Descriptor: | N-oxidanyl-4-[(4-sulfamoylphenyl)methyl]benzamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E65
 
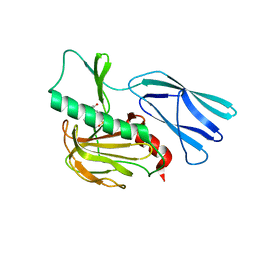 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3 | | Descriptor: | (2S)-2-acetamido-N-[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]-3-(4-sulfamoylphenyl)propanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E64
 
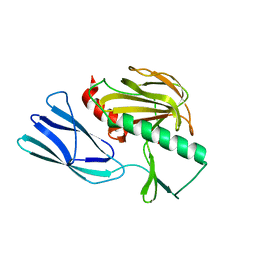 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-2 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E67
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-2 | | Descriptor: | N-oxidanyl-2-[4-(4-sulfamoylphenyl)phenyl]ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E61
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(phenylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Min, K.J, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E63
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-1 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(cyclopentylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E66
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-1 | | Descriptor: | N-[2-(oxidanylamino)-2-oxidanylidene-ethyl]-2-(4-sulfamoylphenyl)ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
4MF2
 
 | | Structure of human DNA polymerase beta complexed with O6MG as the template base in a 1-nucleotide gapped DNA | | Descriptor: | DNA polymerase beta, SODIUM ION, synthetic downstream primer, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
4MFC
 
 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable CTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
4MFF
 
 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable TTP | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
1RZL
 
 | | RICE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN, SULFATE ION | | Authors: | Lee, J.Y, Min, K.S, Cha, H, Shin, D.H, Hwang, K.Y, Suh, S.W. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rice non-specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity.
J.Mol.Biol., 276, 1998
|
|
6MC1
 
 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
6JJL
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-ARG-LYS-LEU, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy.
Commun Biol, 3, 2020
|
|
6JJK
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-TYR-LYS-ILE, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
6JJO
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | Periplasmic serine endoprotease DegP, TMB-CYRKL modulator | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.157 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
