7LIO
 
 | |
7LIN
 
 | |
7LIQ
 
 | |
7LIP
 
 | | X-ray structure of SPOP MATH domain (D140G) | | Descriptor: | SULFATE ION, Speckle-type POZ protein | | Authors: | Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | ATM-phosphorylated SPOP contributes to 53BP1 exclusion from chromatin during DNA replication.
Sci Adv, 7, 2021
|
|
5UMR
 
 | | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1 | | Descriptor: | FACT complex subunit SSRP1 | | Authors: | Su, D, Hu, Q, Thompson, J.R, Heroux, A, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1
To Be Published
|
|
5UMT
 
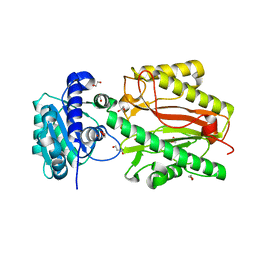 | | Crystal structure of N-terminal domain of human FACT complex subunit SPT16 | | Descriptor: | 1,2-ETHANEDIOL, FACT complex subunit SPT16, GLYCEROL | | Authors: | Su, D, Hu, Q, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SPT16
To Be Published
|
|
5VWE
 
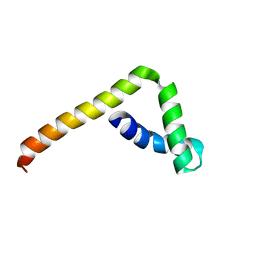 | |
3P8D
 
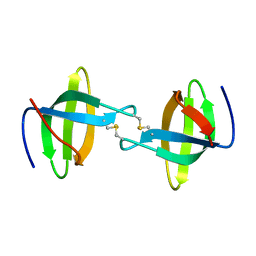 | | Crystal structure of the second Tudor domain of human PHF20 (homodimer form) | | Descriptor: | Medulloblastoma antigen MU-MB-50.72 | | Authors: | Cui, G, Lee, J, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2010-10-13 | | Release date: | 2011-06-22 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2B02
 
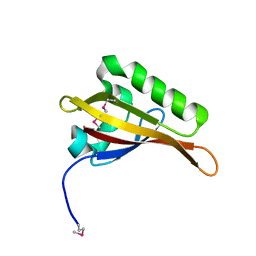 | | Crystal Structure of ARNT PAS-B Domain | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator | | Authors: | Lee, J, Botuyan, M.V, Nomine, Y, Ohh, M, Thompson, J.R, Mer, G. | | Deposit date: | 2005-09-12 | | Release date: | 2006-10-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Binding Properties of ARNT PAS-B Heterodimerization Domain
To be Published
|
|
3PD7
 
 | |
3SD4
 
 | |
2FHD
 
 | | Crystal structure of Crb2 tandem tudor domains | | Descriptor: | DNA repair protein rhp9/CRB2, PHOSPHATE ION | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2005-12-23 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
2ETX
 
 | | Crystal Structure of MDC1 Tandem BRCT Domains | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Wasielewski, E, Kim, Y, Joachimiak, A, Thompson, J.R, Mer, G. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Basis for the Association of Microcephalin (MCPH1) Protein with the Cell Division Cycle Protein 27 (Cdc27) Subunit of the Anaphase-promoting Complex.
J.Biol.Chem., 287, 2012
|
|
3PA6
 
 | |
3Q68
 
 | |
3Q66
 
 | |
2LDM
 
 | |
2LH0
 
 | |
2LVM
 
 | |
2LY1
 
 | |
2MWO
 
 | |
2LH9
 
 | |
2LY2
 
 | |
2L0G
 
 | |
2L0F
 
 | |
