3QXS
 
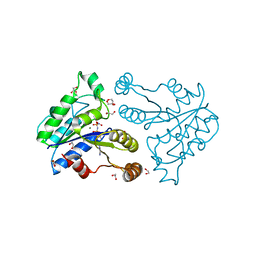 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ANP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, MAGNESIUM ION, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Jablonska, K, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXX
 
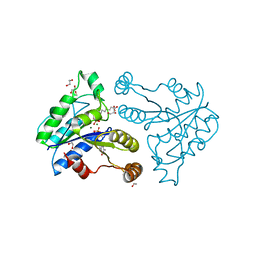 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, Dethiobiotin synthetase, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
4YAB
 
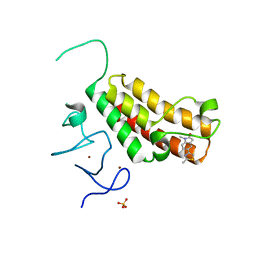 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 1-methyl-5-(2-methyl-1 3-thiazol-4-yl)-2 3-dihydro-1H-indol-2-one (1) | | Descriptor: | 1-methyl-5-(2-methyl-1,3-thiazol-4-yl)-1,3-dihydro-2H-indol-2-one, SULFATE ION, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAD
 
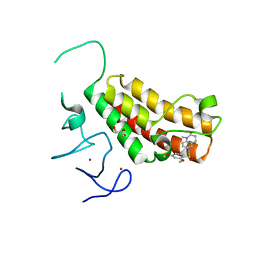 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 2,4-dimethoxy-N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)benzene-1-sulfonamide (3b) | | Descriptor: | 2,4-dimethoxy-N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)benzenesulfonamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBM
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide (7b) | | Descriptor: | GLYCEROL, N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}-3,4-dimethoxybenzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBS
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide (7g) | | Descriptor: | DIMETHYL SULFOXIDE, N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-4-methoxybenzene-1-sulfonamide (5b) | | Descriptor: | N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-4-methoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ZINC ION | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-2-oxo-6-[3-(oxolan-3-ylmethoxy)phenoxy]-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (7l) | | Descriptor: | DIMETHYL SULFOXIDE, N-(1,3-dimethyl-2-oxo-6-{3-[(3S)-tetrahydrofuran-3-ylmethoxy]phenoxy}-2,3-dihydro-1H-benzimidazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YC9
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-3,4-dimethoxybenzene-1-sulfonamide (8i) | | Descriptor: | GLYCEROL, N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-3,4-dimethoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAX
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl]benzenesulfonamide (5g) | | Descriptor: | GLYCEROL, N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
5E1E
 
 | | Human JAK1 kinase in complex with compound 30 at 2.30 Angstroms resolution | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, DI(HYDROXYETHYL)ETHER, Tyrosine-protein kinase JAK1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2015-12-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of azabenzimidazoles as potent JAK1 selective inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2XA4
 
 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-CHLORO-N2-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHYL]-N4-(5-METHYL-1H-PYRAZOL-3-YL)PYRIMIDINE-2,4-DIAMINE, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2010-03-26 | | Release date: | 2010-12-15 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of 5-Chloro-N2-[(1S)-1-(5-Fluoropyrimidin-2-Yl) Ethyl]-N4-(5-Methyl-1H-Pyrazol-3-Yl)Pyrimidine-2,4-Diamine (Azd1480) as a Novel Inhibitor of the Jak/Stat Pathway
J.Med.Chem., 54, 2011
|
|
6RJG
 
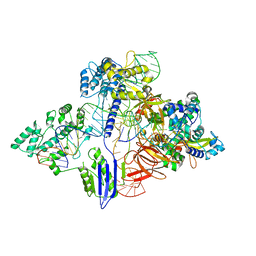 | | Cryo-EM structure of St1Cas9-sgRNA-AcrIIA6-tDNA59-ntPAM complex. | | Descriptor: | AcrIIA6, Cas 9, ntPAM, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJA
 
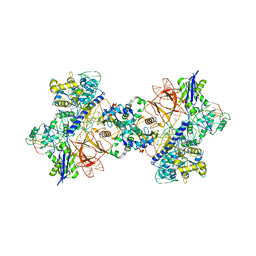 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 dimeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, RNA (78-MER), ... | | Authors: | Goulet, A, Cambillau, C, Chaves-Sanjuan, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJD
 
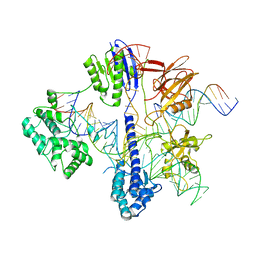 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA59-ntPAM complex. | | Descriptor: | Streptococcus Thermophilus 1 Cas9, ntPAM, sgRNA (78-MER), ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
3ZMM
 
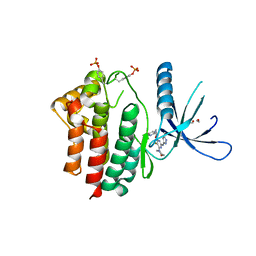 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-FLUORO-4-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHOXY]-N-(5-METHYL-1H-PYRAZOL-3-YL)-6-MORPHOLINO-PYRIMIDIN-2-AMINE, ACETYL GROUP, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Novel Jak2-Stat Pathway Inhibitors with Extended Residence Time on Target.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6RJ9
 
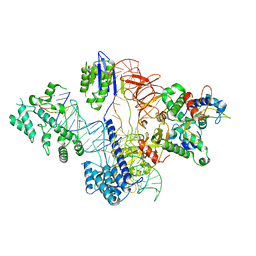 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 monomeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, sgRNA, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
5A70
 
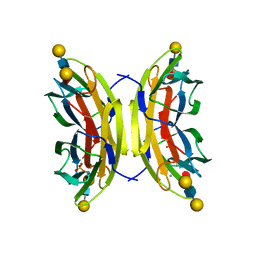 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with lewis x tetrasaccharide | | Descriptor: | CALCIUM ION, LECB, SULFATE ION, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
7OTE
 
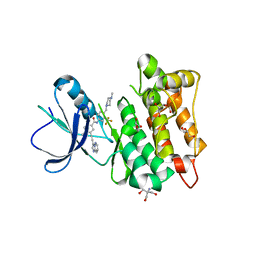 | | Src Kinase Domain in complex with ponatinib | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, GLYCEROL, ... | | Authors: | Soriano-Maldonado, P, Cuesta-Hernandez, H.N, Plaza-Menacho, I. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric switch between the activation loop and a c-terminal palindromic phospho-motif controls c-Src function.
Nat Commun, 14, 2023
|
|
4PLA
 
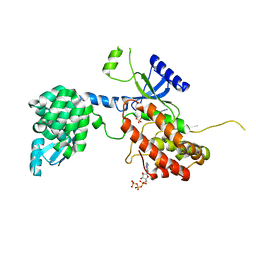 | |
5H8B
 
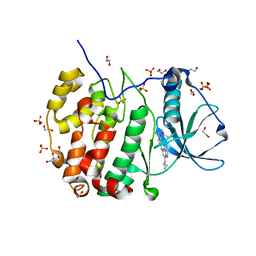 | | Crystal structure of CK2 with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Potent and Selective CK2 Kinase Inhibitors with Effects on Wnt Pathway Signaling in Vivo.
Acs Med.Chem.Lett., 7, 2016
|
|
5H8G
 
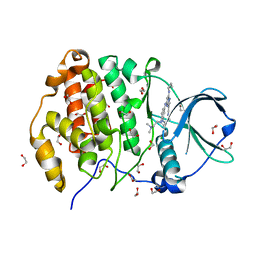 | | Crystal structure of CK2 with compound 7b | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective CK2 Kinase Inhibitors with Effects on Wnt Pathway Signaling in Vivo.
Acs Med.Chem.Lett., 7, 2016
|
|
5H8E
 
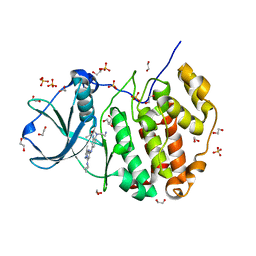 | | Crystal structure of CK2 with compound 7h | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and Selective CK2 Kinase Inhibitors with Effects on Wnt Pathway Signaling in Vivo.
Acs Med.Chem.Lett., 7, 2016
|
|
6QWT
 
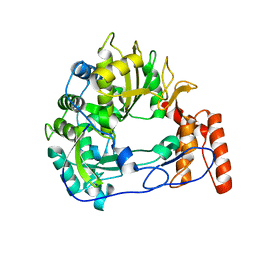 | | Sicinivirus 3Dpol RNA dependent RNA polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
6R1I
 
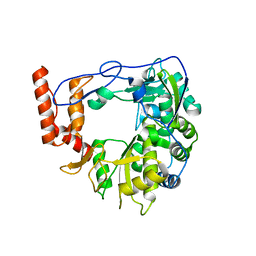 | | Structure of porcine Aichi virus polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
