4GN0
 
 | | De novo phasing of a Hamp-complex using an improved Arcimboldo method | | Descriptor: | Hamp domain of AF1503, MAGNESIUM ION | | Authors: | Hulko, M, Ursinus, A, Bar, K, Martin, J, Zeth, K, Lupas, A.N. | | Deposit date: | 2012-08-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
2DDX
 
 | |
1FHG
 
 | | HIGH RESOLUTION REFINEMENT OF TELOKIN | | Descriptor: | TELOKIN | | Authors: | Tomchick, D.R, Minor, W, Kiyatkin, A, Lewinski, K, Somlyo, A.V, Somlyo, A.P. | | Deposit date: | 2000-08-01 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
7VLK
 
 | | eIF2B-SFSV NSs C2-imposed | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
5URM
 
 | | Crystal structure of human BRR2 in complex with T-1206548 | | Descriptor: | 3-(5-{[(2R)-5-amino-2-cyclohexyl-7-oxo-2,3-dihydro-7H-[1,3,4]thiadiazolo[3,2-a]pyrimidin-6-yl]methyl}furan-2-yl)benzoic acid, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-11 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URJ
 
 | | Crystal structure of human BRR2 in complex with T-3905516 | | Descriptor: | 6-benzyl-3-[(2R)-2-(3-fluoropyridin-2-yl)-6-methyl-3,4-dihydro-2H-1-benzopyran-7-yl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(3H,8H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URK
 
 | | Crystal structure of human BRR2 in complex with T-3935799 | | Descriptor: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Qin, L, Tjhen, R, Klein, M.G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5ZC9
 
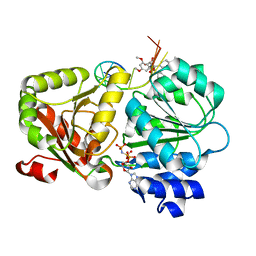 | | Crystal structure of the human eIF4A1-ATP analog-RocA-polypurine RNA complex | | Descriptor: | (1R,2R,3S,3aR,8bS)-6,8-dimethoxy-3a-(4-methoxyphenyl)-N,N-dimethyl-1,8b-bis(oxidanyl)-3-phenyl-2,3-dihydro-1H-cyclopenta[b][1]benzofuran-2-carboxamide, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Takahashi, M, Sakamoto, A, Iwasaki, S, Ito, T. | | Deposit date: | 2018-02-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA.
Mol. Cell, 73, 2019
|
|
6XOG
 
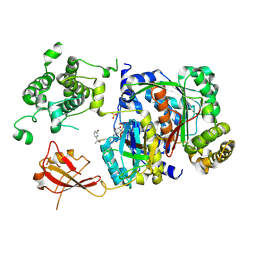 | | Structure of SUMO1-ML786519 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
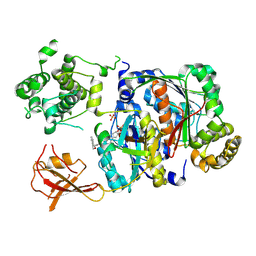 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7F67
 
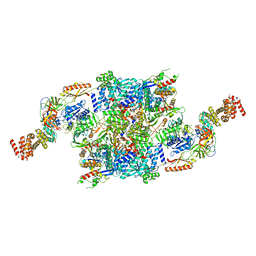 | | eIF2B-SFSV NSs-2-eIF2 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, Non-structural protein NS-S, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7F66
 
 | | eIF2B-SFSV NSs-1-eIF2 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, Non-structural protein NS-S, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7F64
 
 | | eIF2B-SFSV NSs | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7D4C
 
 | | Structure of L-lysine oxidase precursor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, PHOSPHATE ION | | Authors: | Ito, N, Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7D4E
 
 | | Structure of L-lysine oxidase precursor in complex with L-lysine (1.0 M) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, LYSINE | | Authors: | Kitagawa, M, Ito, N, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7D4D
 
 | | Structure of L-lysine oxidase precursor in complex with L-lysine (1.24M) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-Lysine alpha-oxidase, LYSINE | | Authors: | Kitagawa, M, Ito, N, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of enzyme activity regulation by the propeptide of l-lysine alpha-oxidase precursor from Trichoderma viride .
J Struct Biol X, 5, 2021
|
|
7PCM
 
 | | BurG (holo) in complex with (Z)-2,3-dihydroxy-6-methyl-hept-2-enoate (13): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-6-methyl-2,3-bis(oxidanyl)hept-2-enoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCI
 
 | | BurG (holo) in complex with hydroxypyruvate-enol (8): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-2,3-bis(oxidanyl)prop-2-enoic acid, IMIDAZOLE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCL
 
 | | BurG (holo) in complex with 2-hydroxy-2-(hydroxy(isopropyl)amino)acetate (11): Biosynthesis of cyclopropanolrings in bacterial toxins | | Descriptor: | (2S)-2-oxidanyl-2-[oxidanyl(propan-2-yl)amino]ethanoic acid, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCN
 
 | | BurG (holo) in complex with gonyenediol (14), trigonic acid (6) and DMS: Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R)-2-oxidanyl-2-(1-oxidanylcyclopropyl)ethanoic acid, (METHYLSULFANYL)METHANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCE
 
 | | BurG (apo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketol-acid reductoisomerase, PHOSPHATE ION | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCO
 
 | | BurG E232Q mutant (holo) in complex with 2R,3R-2,3-dihydroxy-6-methyl-heptanoate (12): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R,3R)-6-methyl-2,3-bis(oxidanyl)heptanoic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCT
 
 | | BurG E232Q mutant (holo) in complex with enol-oxalacetate (15): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCC
 
 | | BurG in complex with Mg2+ and NAD+ (holo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
