6ZJ6
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with cyclohexylmethyl-Glc-1,3-isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ACETATE ION, ... | | Authors: | Thompson, A.J, Sobala, L.F, Fernandes, P.Z, Hakki, Z, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5C86
 
 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, apo form | | Descriptor: | Kelch domain-containing protein | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
5C92
 
 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, in complex with copper | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (I) ION, ... | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
4UTF
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine and alpha- 1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Cuskin, F, Lowe, E.C, Temple, M.J, Zhu, Y, Pudlo, N.A, Cameron, E.A, Urs, K, Thompson, A.J, Cartmell, A, Rogowski, A, Tolbert, T, Piens, K, Bracke, D, Vervecken, W, Hakki, Z, Speciale, G, Munoz-Munoz, J.L, Pena, M.J, McLean, R, Suits, M.D, Boraston, A.B, Atherly, T, Ziemer, C.J, Williams, S.J, Davies, G.J, Abbott, D.W, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4V1R
 
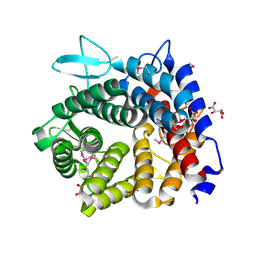 | | Structure of a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, S,R MESO-TARTARIC ACID | | Authors: | Thompson, A.J, Cuskin, F, Spears, R.J, Dabin, J, Turkenburg, J.P, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Gh76 Alpha-Mannanase Homolog, Bt2949, from the Gut Symbiont Bacteroides Thetaiotaomicron
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4V1Z
 
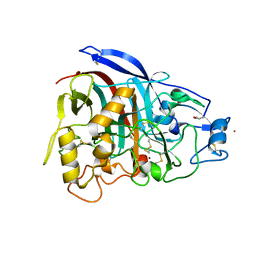 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE, ZINC ION | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8C54
 
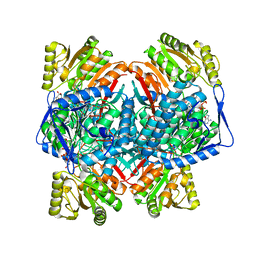 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
3ZMR
 
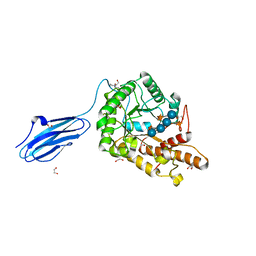 | | Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CELLULASE (GLYCOSYL HYDROLASE FAMILY 5), ... | | Authors: | Larsbrink, J, Rogers, T.E, Hemsworth, G.R, McKee, L.S, Spadiut, O, Klinter, S, Pudlo, N.A, Urs, K, Kelly, A.G, Cederholm, S.N, Davies, G.J, Martens, E.C, Brumer, H. | | Deposit date: | 2013-02-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Discrete Genetic Locus Confers Xyloglucan Metabolism in Select Human Gut Bacteroidetes
Nature, 506, 2014
|
|
8AWR
 
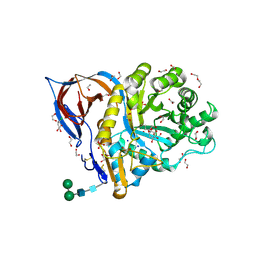 | | Structure of recombinant human beta-glucocerebrosidase in complex with L-carbaxylosyl chloride | | Descriptor: | (1~{S},2~{R},3~{S},6~{S})-6-chloranylcyclohex-4-ene-1,2,3-triol, (1~{S},2~{S},3~{S},4~{R})-cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8AWK
 
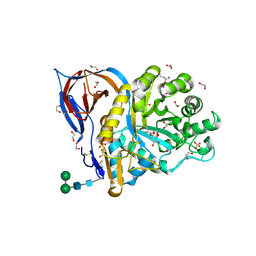 | | Structure of recombinant human beta-glucocerebrosidase in complex with D-carbaxylosyl chloride | | Descriptor: | (2~{S},3~{S},4~{R})-cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8AX3
 
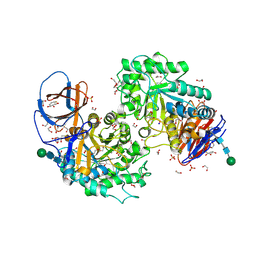 | | Structure of recombinant human beta-glucocerebrosidase in complex with L-carbaxylosyl fluoride | | Descriptor: | (1~{S},2~{R},3~{S},6~{S})-6-fluoranylcyclohex-4-ene-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8BC3
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, BmSF-TAL | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC2
 
 | | Ligand-Free Structure of the decameric sulfofructose transaldolase BmSF-TAL | | Descriptor: | Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC4
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1 | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8B5M
 
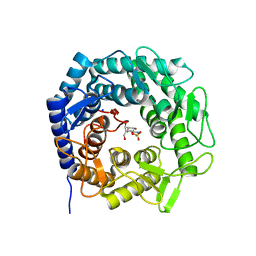 | | Crystal structure of GH47 alpha-1,2-mannosidase from Caulobacter K31 strain in complex with cyclosulfamidate inhibitor | | Descriptor: | (3aR,4S,5S,6R,7R,7aS)-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3a,4,5,6,7,7a-hexahydro-3H-benzo[d][1,2,3]oxathiazole-4,5,6-triol, CALCIUM ION, Mannosyl-oligosaccharide 1,2-alpha-mannosidase, ... | | Authors: | Males, A, Davies, G.J. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | GH47 and Cyclosulfamidate
To Be Published
|
|
8B7P
 
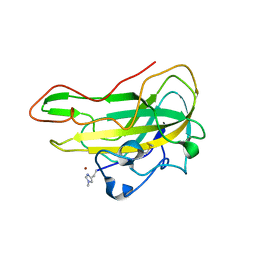 | | Crystal structure of an AA9 LPMO from Aspergillus nidulans, AnLPMOC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Endo-beta-1,4-glucanase D | | Authors: | Males, A, Rafael Fanchini Terrasan, C, Davies, G.J, Walton, P.H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterisation of lytic polysaccharide monooxygenases from Aspergillus nidulans
To Be Published
|
|
8BQA
 
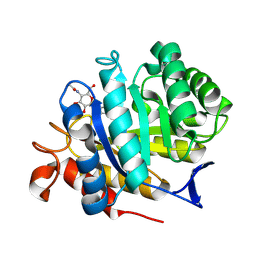 | | CjCel5B endo-glucanase bound to CB665 covalent inhibitor | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, Endoglucanase, alpha-D-xylopyranose, ... | | Authors: | McGregor, N.G.S, de Boer, C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8BN7
 
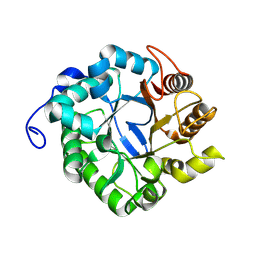 | | CjCel5C endo-glucanase | | Descriptor: | Cellulase, putative, cel5C | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2022-11-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8BQC
 
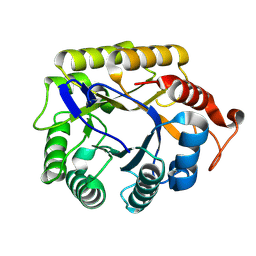 | | CjCel5B endo-glucanase | | Descriptor: | Endoglucanase | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8BQB
 
 | | CjCel5C endo-glucanase bound to CB396 covalent inhibitor | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, Cellulase, putative, ... | | Authors: | McGregor, N.G.S, de Boer, C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8C4I
 
 | |
8CE3
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with 3D fragment 2548 | | Descriptor: | (3~{S})-1-ethanoyl-3-(4-methylphenyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Gilio, A.K, Darby, J.F, Wu, L, Davies, G.J. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Modular 3-D Fragment Collection: Design, Synthesis and Screening
To Be Published
|
|
8CM9
 
 | | Structure of human O-GlcNAc transferase in complex with UDP and tP11 | | Descriptor: | PHE-MET-PRO-LYS-TYR-SER-ILE, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2023-02-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7DIF
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol at 1.75-angstrom resolution | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, POTASSIUM ION, ... | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5IJU
 
 | | Structure of an AA10 Lytic Polysaccharide Monooxygenase from Bacillus amyloliquefaciens with Cu(II) bound | | Descriptor: | 1,2-ETHANEDIOL, BaAA10 Lytic Polysaccharide Monooxygenase, CALCIUM ION, ... | | Authors: | Gregory, R.C, Hemsworth, G.R, Turkenburg, J.P, Hart, S.J, Walton, P.H, Davies, G.J. | | Deposit date: | 2016-03-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity, stability and 3-D structure of the Cu(ii) form of a chitin-active lytic polysaccharide monooxygenase from Bacillus amyloliquefaciens.
Dalton Trans, 45, 2016
|
|
