5XZW
 
 | | Crystal structure of Rad53 1-466 | | Descriptor: | Serine/threonine-protein kinase RAD53 | | Authors: | Weng, J.H, Tsai, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phospho-Priming Confers Functionally Relevant Specificities for Rad53 Kinase Autophosphorylation
Biochemistry, 56, 2017
|
|
5XZV
 
 | | Crystal structure of Rad53 1-466 in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase RAD53 | | Authors: | Weng, J.H, Tsai, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phospho-Priming Confers Functionally Relevant Specificities for Rad53 Kinase Autophosphorylation
Biochemistry, 56, 2017
|
|
7V29
 
 | | Crystal structure of FGFR4 with a dual-warhead covalent inhhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[3-(3,5-dimethoxyphenyl)-2-oxidanylidene-1-[3-(4-propanoylpiperazin-1-yl)propyl]-4H-pyrimido[4,5-d]pyrimidin-7-yl]amino]phenyl]propanamide, SULFATE ION | | Authors: | Chen, X.J, Jiang, L.Y, Dai, S.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-08-07 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Structure-based design of a dual-warhead covalent inhibitor of FGFR4.
Commun Chem, 5, 2022
|
|
8YMB
 
 | | The crystal structure of SHD931 in complex with Brd4-BD2 and VCB | | Descriptor: | Bromodomain-containing protein 4, CHLORIDE ION, Elongin-B, ... | | Authors: | Huang, W.X, Chen, Y.H, Li, C.G, Ding, K. | | Deposit date: | 2024-03-08 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Based Design of "Head-to-Tail" Macrocyclic PROTACs.
Jacs Au, 4, 2024
|
|
7Y6T
 
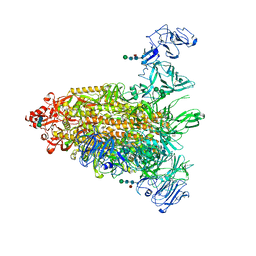 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6U
 
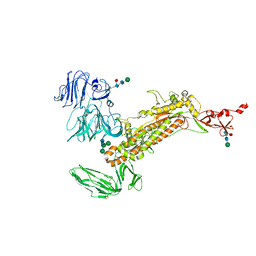 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6V
 
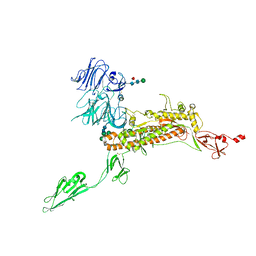 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6S
 
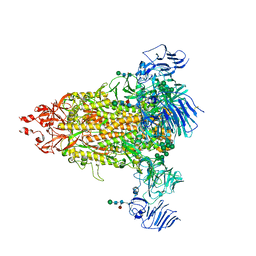 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7W73
 
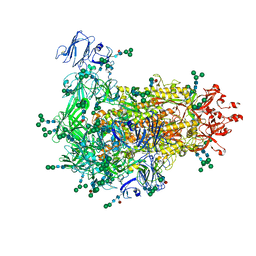 | | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-03 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7W6M
 
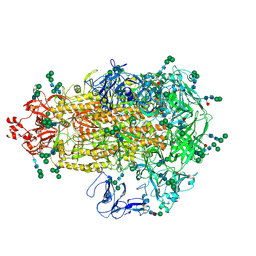 | | Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
