6B4Z
 
 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase, T157S Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein | | Authors: | Taylor, A.B. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Why does oxamniquine kill Schistosoma mansoni and not S. haematobium and S. japonicum?
Int.J.Parasitol., 2020
|
|
6CJ7
 
 | | Crystal structure of Manduca sexta Serine protease inhibitor (Serpin)-12 | | Descriptor: | Serpin-12 | | Authors: | Gulati, M, Hu, Y, Peng, S, Pathak, P.K, Wang, Y, Deng, J, Jiang, H. | | Deposit date: | 2018-02-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Manduca sexta serpin-12 controls the prophenoloxidase activation system in larval hemolymph.
Insect Biochem. Mol. Biol., 99, 2018
|
|
7XJZ
 
 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA binding state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XJY
 
 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase (apo form) | | Descriptor: | Glycine--tRNA ligase | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK1
 
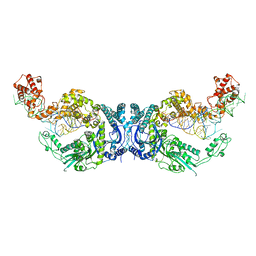 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (both in tRNA binding states) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK0
 
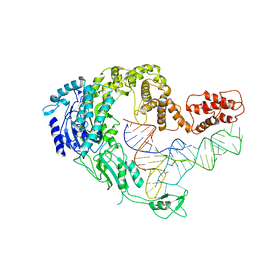 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA locked state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
2KCJ
 
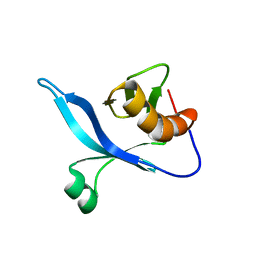 | | solution structure of FAPP1 PH domain | | Descriptor: | Pleckstrin homology domain-containing family A member 3 | | Authors: | Lenoir, M, Coskun, U, James, J, Simons, K, Overduin, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains.
Embo Rep., 11, 2010
|
|
2NNX
 
 | |
3R0Q
 
 | |
5GNG
 
 | |
5H3B
 
 | |
8X6B
 
 | | Crystal structure of immune receptor PVRIG in complex with ligand Nectin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nectin-2, Transmembrane protein PVRIG | | Authors: | Hu, S.T, Han, P, Wang, H, Qi, J.X. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Structure, 32, 2024
|
|
8H2X
 
 | | Structure of Acb2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H39
 
 | | Structure of Acb2 complexed with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H2J
 
 | | Structure of Acb2 complexed with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
6IP0
 
 | |
6IP4
 
 | |
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
8G05
 
 | | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | | Descriptor: | 6-(octylamino)pyrimidine-2,4(3H,5H)-dione, CHOLESTEROL, G-protein coupled receptor 84, ... | | Authors: | Zhang, X, Wang, Y.J, Li, X, Liu, G.B, Gong, W.M, Zhang, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Pro-phagocytic function and structural basis of GPR84 signaling.
Nat Commun, 14, 2023
|
|
6KXG
 
 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
5YKO
 
 | |
7E4I
 
 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40/Tom5/Tom6 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, Mitochondrial import receptor subunit TOM6, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
7E4H
 
 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
5YKN
 
 | | crystal structure of Arabidopsis thaliana JMJ14 catalytic domain | | Descriptor: | NICKEL (II) ION, Probable lysine-specific demethylase JMJ14, ZINC ION | | Authors: | Yang, Z, Du, J. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases.
Plant Cell, 30, 2018
|
|
6JNL
 
 | | REF6 ZnF2-4-NAC004 complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
