5D6L
 
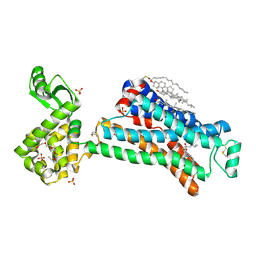 | | beta2AR-T4L - CIM | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Ma, P, Caffrey, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The cubicon method for concentrating membrane proteins in the cubic mesophase.
Nat Protoc, 12, 2017
|
|
7TP3
 
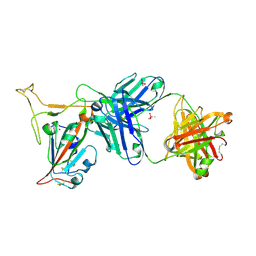 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | Descriptor: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
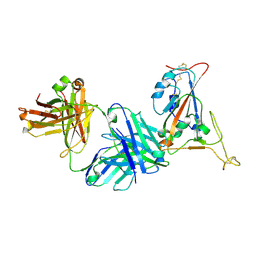 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
1WKO
 
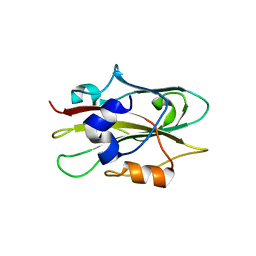 | |
1WKP
 
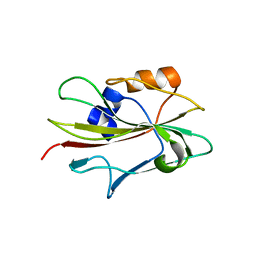 | | Flowering locus t (ft) from arabidopsis thaliana | | Descriptor: | FLOWERING LOCUS T protein, SULFATE ION | | Authors: | Miller, D, Banfield, M.J, Winter, V.J, Brady, R.L. | | Deposit date: | 2004-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A divergent external loop confers antagonistic activity on floral regulators FT and TFL1.
Embo J., 25, 2006
|
|
7NE3
 
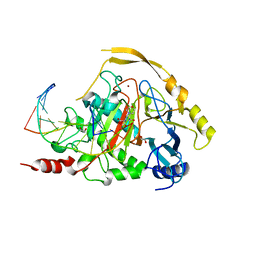 | | Human TET2 in complex with favourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
7NE6
 
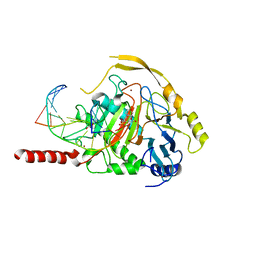 | | Human TET2 in complex with unfavourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
6SWY
 
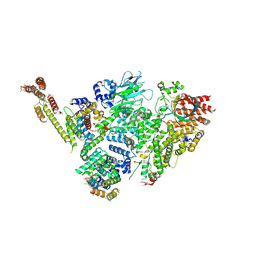 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | Descriptor: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
6NE8
 
 | |
5EQW
 
 | | Structure of the major structural protein D135 of Acidianus tailed spindle virus (ATSV) | | Descriptor: | NITRATE ION, Putative major coat protein | | Authors: | Hochstein, R.A, Lintner, N.G, Young, M.J, Lawrence, C.M. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-16 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Structural studies ofAcidianustailed spindle virus reveal a structural paradigm used in the assembly of spindle-shaped viruses.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6YBQ
 
 | | Engineered glycolyl-CoA carboxylase (quintuple mutant) with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
6YBP
 
 | | Propionyl-CoA carboxylase of Methylorubrum extorquens with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
6UDK
 
 | | HIV-1 bNAb 1-55 in complex with modified BG505 SOSIP-based immunogen RC1 and 10-1074 | | Descriptor: | 1-55 Fab Heavy Chain, 1-55 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6UDJ
 
 | | HIV-1 bNAb 1-18 in complex with BG505 SOSIP.664 and 10-1074 | | Descriptor: | 1-18 Fab Heavy Chain, 1-18 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
7B0O
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus in space group P21 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7B0P
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus in space group P21212 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, GLYCEROL, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7B0Q
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus with H85A mutation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CITRIC ACID, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7B0R
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus with H85R mutation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7ACI
 
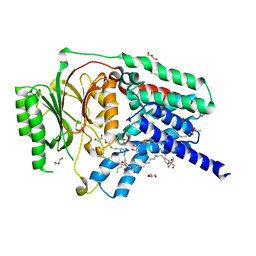 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 9.8 monoacylglycerol | | Descriptor: | Apolipoprotein N-acyltransferase, GLYCEROL, [(2~{S})-2,3-bis(oxidanyl)propyl] heptadec-9-enoate | | Authors: | Smithers, L, van Dalsen, L, Boland, C, Caffrey, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins
Cryst.Growth Des., 2020
|
|
7ACG
 
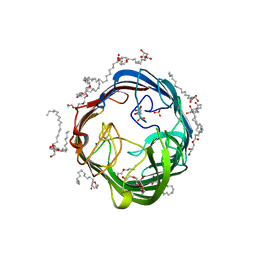 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 9.8 monoacylglycerol | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-(2-METHOXYETHOXY)ETHANOL, Alginate biosynthesis protein AlgE, ... | | Authors: | Smithers, L, van Dalsen, L, Boland, C, Caffrey, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins
Cryst.Growth Des., 2020
|
|
8B0K
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form) | | Descriptor: | Apolipoprotein N-acyltransferase | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0P
 
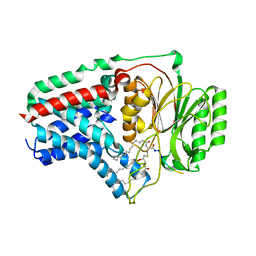 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3 | | Descriptor: | Apolipoprotein N-acyltransferase, Pam3-SKKKK, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0O
 
 | | Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3 | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{R})-3-[(2~{R})-3-[[(2~{R})-1-[[(2~{R})-1-[[(2~{R})-6-[(2-aminophenyl)carbonylamino]-1-azanyl-1-oxidanylidene-hexan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0L
 
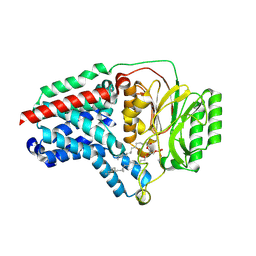 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0M
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant) | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
