3VRB
 
 | | Mitochondrial rhodoquinol-fumarate reductase from the parasitic nematode Ascaris suum with the specific inhibitor flutolanil and substrate fumarate | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Shimizu, H, Shiba, T, Inaoka, D.K, Osanai, A, Kita, K, Sakamoto, K, Harada, S. | | Deposit date: | 2012-04-07 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of mitochondrial quinol-fumarate reductase from the parasitic nematode Ascaris suum
J.Biochem., 151, 2012
|
|
1WTE
 
 | | Crystal structure of type II restrcition endonuclease, EcoO109I complexed with cognate DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*GP*CP*CP*CP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*GP*GP*GP*CP*CP*CP*GP*GP*T)-3', EcoO109IR, ... | | Authors: | Hashimoto, H, Shimizu, T, Imasaki, T, Kato, M, Shichijo, N, Kita, K, Sato, M. | | Deposit date: | 2004-11-22 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of type II restriction endonuclease EcoO109I and its complex with cognate DNA
J.Biol.Chem., 280, 2005
|
|
1WTD
 
 | | Crystal structure of type II restrcition endonuclease, EcoO109I DNA-free form | | Descriptor: | EcoO109IR, GLYCEROL | | Authors: | Hashimoto, H, Shimizu, T, Imasaki, T, Kato, M, Shichijo, N, Kita, K, Sato, M. | | Deposit date: | 2004-11-22 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structures of type II restriction endonuclease EcoO109I and its complex with cognate DNA
J.Biol.Chem., 280, 2005
|
|
1UMG
 
 | | Crystal structure of fructose-1,6-bisphosphatase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), 385aa long conserved hypothetical protein, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-09-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
Structure, 12, 2004
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
2ZHV
 
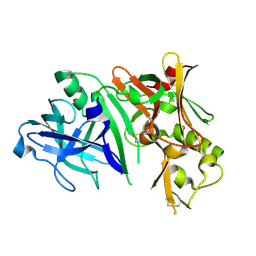 | | Crystal structure of BACE1 at pH 7.0 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZHS
 
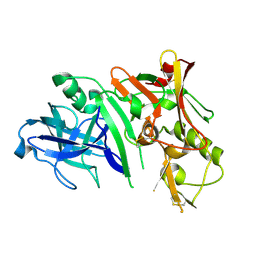 | | Crystal structure of BACE1 at pH 4.0 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZHU
 
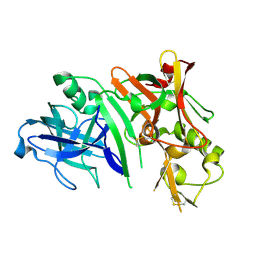 | | Crystal structure of BACE1 at pH 5.0 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZO0
 
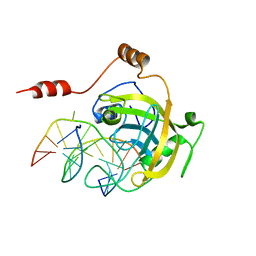 | | mouse NP95 SRA domain DNA specific complex 1 | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
2ZHT
 
 | | Crystal structure of BACE1 at pH 4.5 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZHR
 
 | |
2ZO1
 
 | | Mouse NP95 SRA domain DNA specific complex 2 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), ... | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
2ZO2
 
 | | Mouse NP95 SRA domain non-specific DNA complex | | Descriptor: | DNA (5'-D(*DAP*DAP*DCP*DTP*DGP*DCP*DGP*DCP*DAP*DGP*DTP*DT)-3'), E3 ubiquitin-protein ligase UHRF1, PHOSPHATE ION | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
3AYU
 
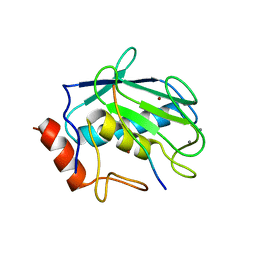 | | Crystal structure of MMP-2 active site mutant in complex with APP-drived decapeptide inhibitor | | Descriptor: | 72 kDa type IV collagenase, Amyloid beta A4 protein, CALCIUM ION, ... | | Authors: | Hashimoto, H, Takeuchi, T, Komatsu, K, Miyazaki, K, Sato, M, Higashi, S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for matrix metalloproteinase-2 (MMP-2)-selective inhibitory action of {beta}-amyloid precursor protein-derived inhibitor
J.Biol.Chem., 2011
|
|
1WNS
 
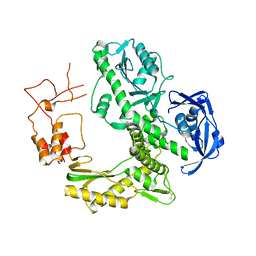 | | Crystal structure of family B DNA polymerase from hyperthermophilic archaeon pyrococcus kodakaraensis KOD1 | | Descriptor: | DNA POLYMERASE | | Authors: | Hashimoto, H, Inoue, T, Kai, Y, Fujiwara, S, Takagi, M, Nishioka, M, Imanaka, T. | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-17 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DNA Polymerase from Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
J.Mol.Biol., 306, 2001
|
|
3W1E
 
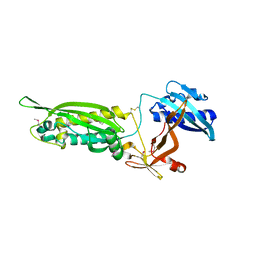 | | Structure of FlgT, a flagellar basal body component protein | | Descriptor: | Flagella basal-body protein | | Authors: | Terashima, H, Sakuma, M, Homma, M, Imada, K. | | Deposit date: | 2012-11-14 | | Release date: | 2013-04-03 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2COV
 
 | | Crystal structure of CBM31 from beta-1,3-xylanase | | Descriptor: | beta-1,3-xylanase | | Authors: | Hashimoto, H, Tamai, Y, Okazaki, F, Tamaru, Y, Shimizu, T, Araki, T, Sato, M. | | Deposit date: | 2005-05-18 | | Release date: | 2005-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The first crystal structure of a family 31 carbohydrate-binding module with affinity to beta-1,3-xylan
Febs Lett., 579, 2005
|
|
2E2P
 
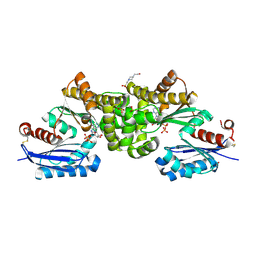 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2E2Q
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with xylose, Mg2+, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE, MAGNESIUM ION, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2E2N
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in the apo form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXOKINASE, SULFATE ION | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2E2O
 
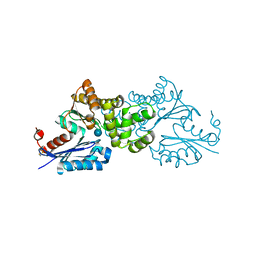 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with glucose | | Descriptor: | HEXOKINASE, beta-D-glucopyranose | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2RSO
 
 | |
2RSN
 
 | |
3A57
 
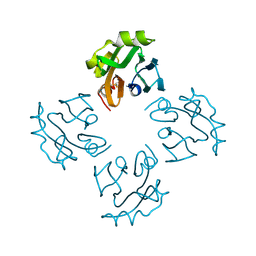 | | Crystal structure of Thermostable Direct Hemolysin | | Descriptor: | Thermostable direct hemolysin 2 | | Authors: | Hashimoto, H, Yanagihara, I, Nakahira, K, Hamada, D, Ikegami, T, Mayanagi, K, Kaieda, S, Fukui, T, Ohnishi, K, Kajiyama, S, Yamane, T, Ikeguchi, M, Honda, T, Shimizu, T, Sato, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin
J.Biol.Chem., 285, 2010
|
|
1F26
 
 | |
