4K97
 
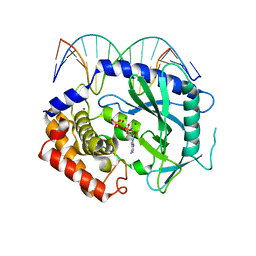 | | Structure of Ternary Complex of cGAS with dsDNA and Bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, DNA-F, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
4NTI
 
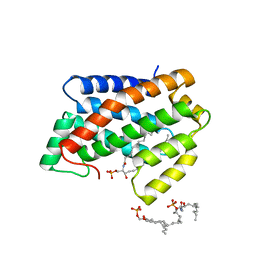 | | Crystal structure of D60N mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.9 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NTG
 
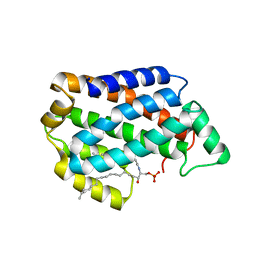 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.55 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5505 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
7TVE
 
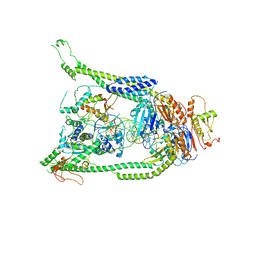 | | ATP and DNA bound SMC5/6 core complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (68-MER), DNA (78-MER), ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
149D
 
 | |
1SLO
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1SLP
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, 16 STRUCTURES | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
7VKJ
 
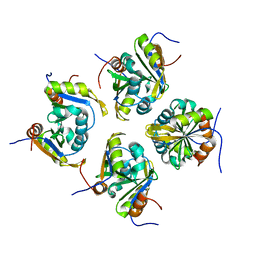 | | Structure of ESRP1 qRRM3 domain | | Descriptor: | Epithelial splicing regulatory protein 1 | | Authors: | Wu, B.X, Patel, D.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of ESRP1 qRRM3 domain
To Be Published
|
|
3VRS
 
 | | Crystal structure of fluoride riboswitch, soaked in Mn2+ | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MANGANESE (II) ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
484D
 
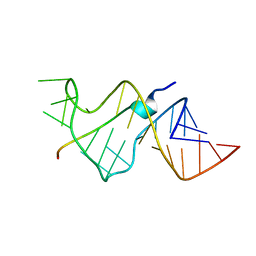 | | SOLUTION STRUCTURE OF HIV-1 REV PEPTIDE-RNA APTAMER COMPLEX | | Descriptor: | BASIC REV PEPTIDE, RNA APTAMER | | Authors: | Ye, X, Gorin, A.A, Frederick, R, Hu, W, Majumdar, A, Xu, W, Mclendon, G, Ellington, A, Patel, D.J. | | Deposit date: | 1999-08-02 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | RNA architecture dictates the conformations of a bound peptide.
Chem.Biol., 6, 1999
|
|
1PIK
 
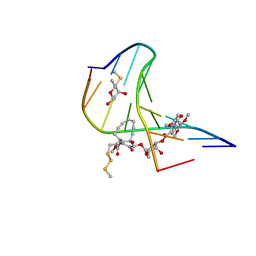 | | ESPERAMICIN A1-DNA COMPLEX, NMR, 4 STRUCTURES | | Descriptor: | 2,4-dideoxy-3-O-methyl-4-(propan-2-ylamino)-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-S-methyl-4-thio-beta-D-ribo-hexopyranose, 2-deoxy-alpha-L-fucopyranose, ... | | Authors: | Kumar, R.A, Ikemoto, N, Patel, D.J. | | Deposit date: | 1996-12-11 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Esperamicin A1-DNA Complex
J.Mol.Biol., 265, 1997
|
|
7R76
 
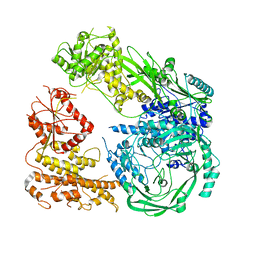 | | Cryo-EM structure of DNMT5 in apo state | | Descriptor: | DNA repair protein Rad8, ZINC ION | | Authors: | Wang, J, Patel, D.J. | | Deposit date: | 2021-06-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
7SDE
 
 | | Cryo-EM structure of Nse5/6 heterodimer | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of Nse5/6 complex with the C terminal part of Nse5
To Be Published
|
|
7SHP
 
 | | Crystal structure of hSTING in complex with c[2',3'-(ribo-2'-G, xylo-3'-A)-MP](RJ244) | | Descriptor: | (2S,5R,7R,8R,10S,12aR,14R,15R,15aR,16R)-7-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-2,10,15,16-tetrahydroxyoctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Xie, W, Lama, L, Yang, X.J, Kuryavyi, V, Nudelman, I, Glickman, J.F, Jones, R.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arabinose- and xylose-modified analogs of 2',3'-cGAMP act as STING agonists.
Cell Chem Biol, 2023
|
|
7SHO
 
 | | Crystal structure of hSTING in complex with c[2',3'-(ara-2'-G, ribo-3'-A)-MP] (RJ242) | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-2,10,15,16-tetrahydroxyoctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Xie, W, Lama, L, Yang, X.J, Kuryavyi, V, Nudelman, I, Glickman, J.F, Jones, R.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2021-10-10 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Arabinose- and xylose-modified analogs of 2',3'-cGAMP act as STING agonists.
Cell Chem Biol, 2023
|
|
8T8E
 
 | | cryoEM structure of Smc5/6 5mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 6 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T8F
 
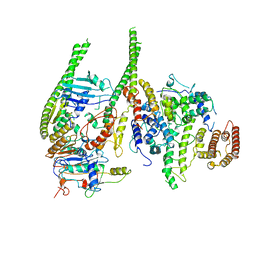 | | Smc5/6 8mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosome element 5, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T66
 
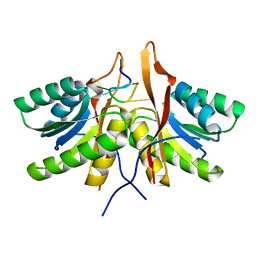 | | cA6 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T65
 
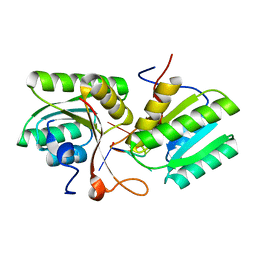 | | cA4 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T64
 
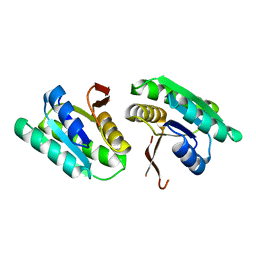 | | Apo Cam1(42-206) | | Descriptor: | Cam1 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8URU
 
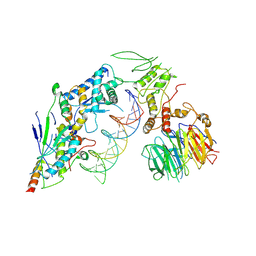 | | Spo11 core complex with hairpin DNA | | Descriptor: | Antiviral protein SKI8, Hairpin DNA, MAGNESIUM ION, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 2024
|
|
8URQ
 
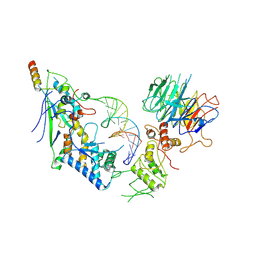 | | Spo11 core complex with gapped DNA | | Descriptor: | Antiviral protein SKI8, MAGNESIUM ION, Meiosis-specific protein SPO11, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 2024
|
|
5ELH
 
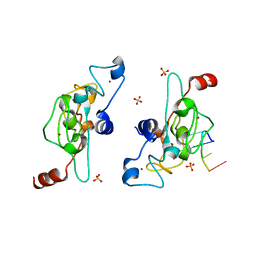 | | Crystal structure of mouse Unkempt zinc fingers 1-3 (ZnF1-3), bound to RNA | | Descriptor: | RING finger protein unkempt homolog, RNA (5'-R(*UP*UP*AP*UP*U)-3'), SULFATE ION, ... | | Authors: | Teplova, M, Murn, J, Zarnack, K, Shi, Y, Patel, D.J. | | Deposit date: | 2015-11-04 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7R77
 
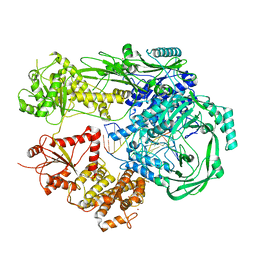 | |
7R78
 
 | |
