4UV7
 
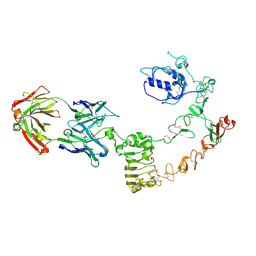 | | The complex structure of extracellular domain of EGFR and GC1118A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Yoo, J.H, Cho, H.S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gc1118, an Anti-Egfr Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity Against High-Affinity Egfr Ligands.
Mol.Cancer Ther., 15, 2016
|
|
6Q21
 
 | |
1IM5
 
 | |
1S7O
 
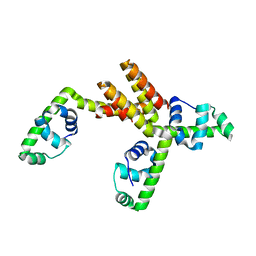 | | Crystal structure of putative DNA binding protein SP_1288 from Streptococcus pygenes | | Descriptor: | Hypothetical UPF0122 protein SPy1201/SpyM3_0842/SPs1042/spyM18_1152 | | Authors: | Oganesyan, V, Pufan, R, DeGiovanni, A, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-29 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the putative DNA-binding protein SP_1288 from Streptococcus pyogenes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1S12
 
 | | Crystal structure of TM1457 | | Descriptor: | ACETATE ION, hypothetical protein TM1457 | | Authors: | Shin, D.H, Lou, Y, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TM1457 from Thermotoga maritima.
J.Struct.Biol., 152, 2005
|
|
1S6X
 
 | | Solution structure of VSTx | | Descriptor: | KvAP CHANNEL | | Authors: | Jung, H.J, Eu, Y.J, Kim, J.I. | | Deposit date: | 2004-01-28 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and lipid membrane partitioning of VSTx1, an inhibitor of the KvAP potassium channel.
Biochemistry, 44, 2005
|
|
8GTG
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-I-152 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N},~{N}-bis(2-methoxyethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTI
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C205 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N}-butyl-~{N}-(cyclopropylmethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1, ... | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTM
 
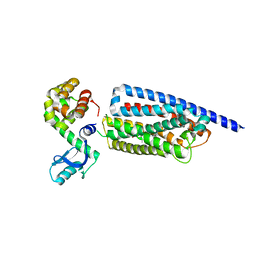 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
2G97
 
 | |
2G96
 
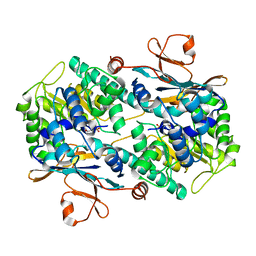 | |
2G95
 
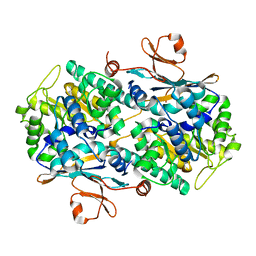 | |
2GQT
 
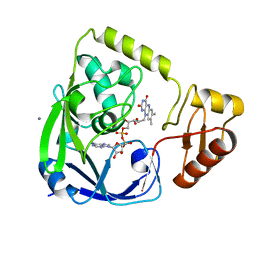 | |
2GQU
 
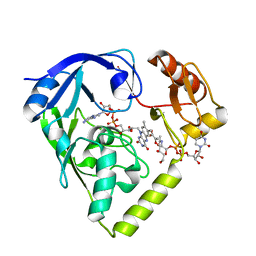 | |
3CO9
 
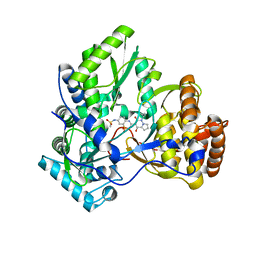 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[4-hydroxy-1-(3-methylbutyl)-2-oxo-1,2-dihydropyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7 -yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-03-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6TNA
 
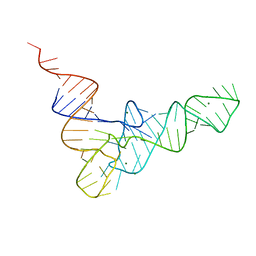 | | CRYSTAL STRUCTURE OF YEAST PHENYLALANINE T-RNA. I.CRYSTALLOGRAPHIC REFINEMENT | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Sussman, J.L, Holbrook, S.R, Warrant, R.W, Church, G.M, Kim, S.-H. | | Deposit date: | 1978-11-16 | | Release date: | 1979-01-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement.
J.Mol.Biol., 123, 1978
|
|
6A6K
 
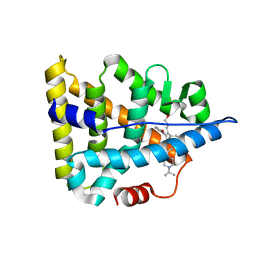 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
1VLS
 
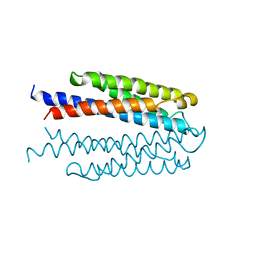 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR | | Descriptor: | ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
1VLT
 
 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR WITH ASPARTATE | | Descriptor: | ASPARTATE RECEPTOR, ASPARTIC ACID | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
1HCL
 
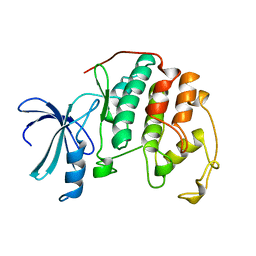 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
1DM2
 
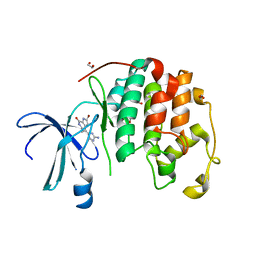 | | HUMAN CYCLIN-DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR HYMENIALDISINE | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Thunnissen, A.M, Kim, S.-H. | | Deposit date: | 1999-12-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of cyclin-dependent kinases, GSK-3beta and CK1 by hymenialdisine, a marine sponge constituent.
Chem.Biol., 7, 2000
|
|
2PYY
 
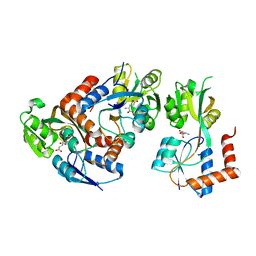 | | Crystal Structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with (L)-glutamate | | Descriptor: | GLUTAMIC ACID, Ionotropic glutamate receptor bacterial homologue | | Authors: | Lee, J.H, Kang, G.B, Lim, H.-H, Ree, M, Park, C.-S, Eom, S.H. | | Deposit date: | 2007-05-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with L-glutamate: structural dissection of the ligand interaction and subunit interface.
J.Mol.Biol., 376, 2008
|
|
1HCK
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HUMAN CYCLIN-DEPENDENT KINASE 2, MAGNESIUM ION | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
6L33
 
 | |
1COJ
 
 | | FE-SOD FROM AQUIFEX PYROPHILUS, A HYPERTHERMOPHILIC BACTERIUM | | Descriptor: | FE (III) ION, PROTEIN (SUPEROXIDE DISMUTASE) | | Authors: | Lim, J.H, Yu, Y.G, Kim, S.-H, Cho, S.-J, Ahn, B.Y, Han, Y.S, Cho, Y. | | Deposit date: | 1999-05-28 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an Fe-superoxide dismutase from the hyperthermophile Aquifex pyrophilus at 1.9 A resolution: structural basis for thermostability.
J.Mol.Biol., 270, 1997
|
|
