7WS5
 
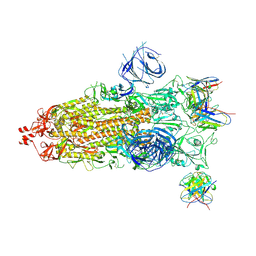 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS1
 
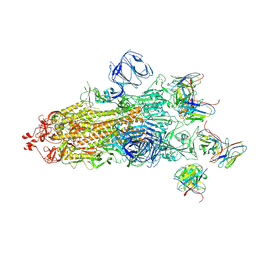 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7X1G
 
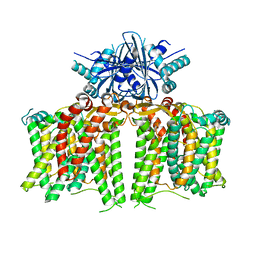 | |
7X1I
 
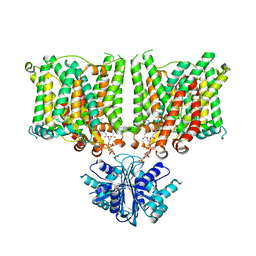 | | Cryo-EM structure of human BTR1 in the outward-facing state. | | Descriptor: | Isoform 1 of Solute carrier family 4 member 11, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y, Lu, Y, Zuo, P. | | Deposit date: | 2022-02-24 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP 2.
Nat Commun, 14, 2023
|
|
7X1H
 
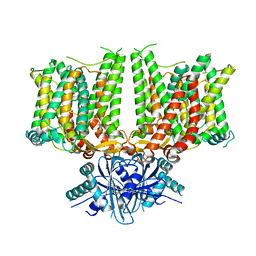 | |
7X1J
 
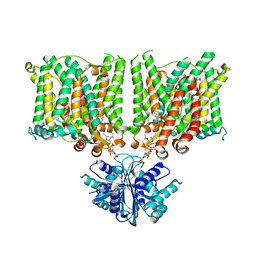 | | Cryo-EM structure of human BTR1 in the outward-facing state in the presence of NH4Cl. | | Descriptor: | Isoform 1 of Solute carrier family 4 member 11, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y, Lu, Y, Zuo, P. | | Deposit date: | 2022-02-24 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP 2.
Nat Commun, 14, 2023
|
|
7XFA
 
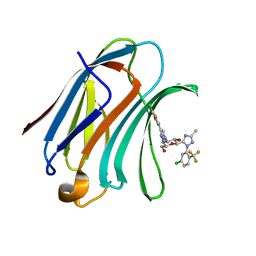 | | Structure of human Galectin-3 CRD in complex with monosaccharide inhibitor | | Descriptor: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[2-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Shukla, J, Raman, S, Ghosh, K. | | Deposit date: | 2022-04-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Identification of Monosaccharide Derivatives as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3.
J.Med.Chem., 65, 2022
|
|
7XM0
 
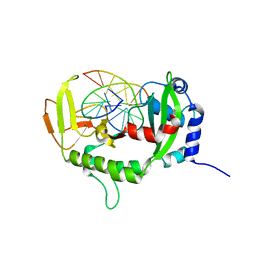 | | Crystal structure of Sau3AI-C and DNA substrate complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*CP*AP*TP*G)-3'), MAGNESIUM ION, Type II restriction enzyme Sau3AI | | Authors: | Liu, Y, Yu, F, He, J. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of Sau3AI with and without bound DNA suggest a self-activation-based DNA cleavage mechanism.
Structure, 31, 2023
|
|
7W7X
 
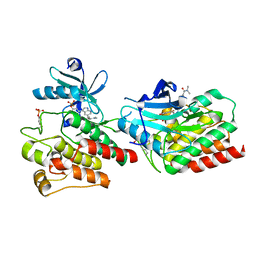 | | The crystal structure of human abl1 kinase domain in complex with ABL1-A11 | | Descriptor: | 5-[5-(dimethylcarbamoyl)pyridin-3-yl]-3-(5-fluorosulfonyloxy-2-methoxy-phenyl)-1H-pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z.M. | | Deposit date: | 2021-12-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.0000093 Å) | | Cite: | Cell-Active, Reversible, and Irreversible Covalent Inhibitors That Selectively Target the Catalytic Lysine of BCR-ABL Kinase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7W9M
 
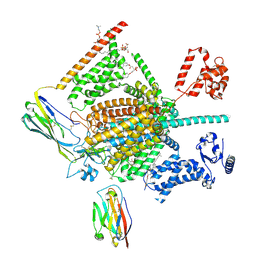 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (S6IV pi helix conformer) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9P
 
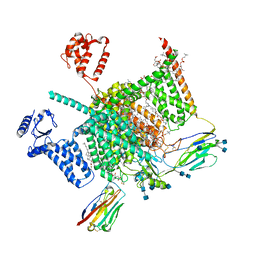 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV pi helix conformer) | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9T
 
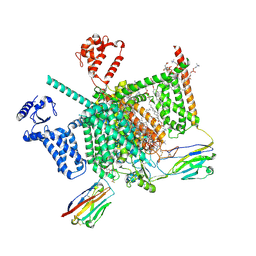 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV alpha helix conformer) | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9L
 
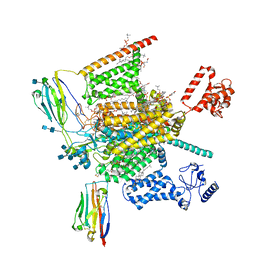 | | Cryo-EM structure of human Nav1.7(E406K)-beta1-beta2 complex | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W1Y
 
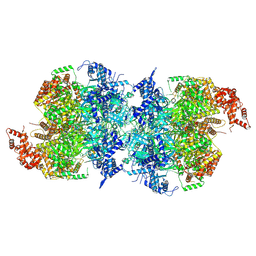 | | Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (49-MER), ... | | Authors: | Li, J, Dong, J, Dang, S, Zhai, Y. | | Deposit date: | 2021-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | The human pre-replication complex is an open complex.
Cell, 186, 2023
|
|
7W7Y
 
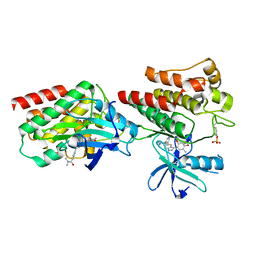 | | The crystal structure of human abl1 kinase domain in complex with ABL2-A5 | | Descriptor: | 5-[3-(5-methanoyl-2-methoxy-4-oxidanyl-phenyl)-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-~{N},~{N}-dimethyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z.M. | | Deposit date: | 2021-12-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20003033 Å) | | Cite: | Cell-Active, Reversible, and Irreversible Covalent Inhibitors That Selectively Target the Catalytic Lysine of BCR-ABL Kinase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7C4A
 
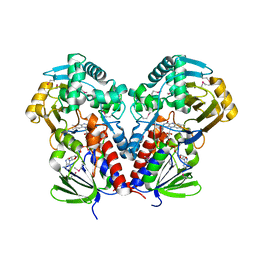 | | nicA2 with cofactor FAD | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
7C49
 
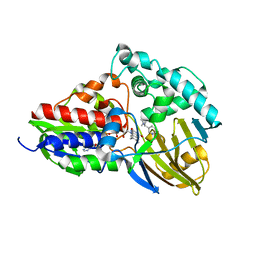 | | nicA2 with cofactor FAD and substrate nicotine | | Descriptor: | 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zhang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
7DNU
 
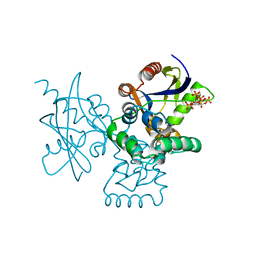 | | mRNA-decapping enzyme g5Rp with inhibitor insp6 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7DWQ
 
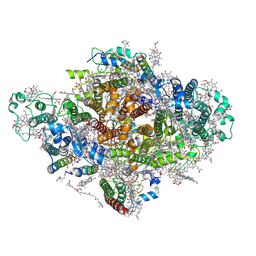 | | Photosystem I from a chlorophyll d-containing cyanobacterium Acaryochloris marina | | Descriptor: | (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Chen, J.H, Zhang, X, Shen, J.R. | | Deposit date: | 2021-01-17 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A unique photosystem I reaction center from a chlorophyll d-containing cyanobacterium Acaryochloris marina.
J Integr Plant Biol, 63, 2021
|
|
7DXH
 
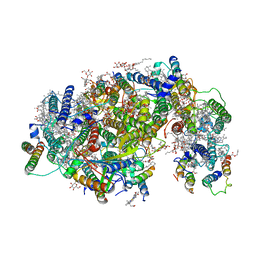 | | Cryo-EM structure of PSII intermediate Psb28-PSII complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Sui, S.F, Shen, J.R, Han, G.Y, Xiao, Y.N, Huang, G.Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into cyanobacterial photosystem II intermediates associated with Psb28 and Tsl0063.
Nat.Plants, 7, 2021
|
|
7DXA
 
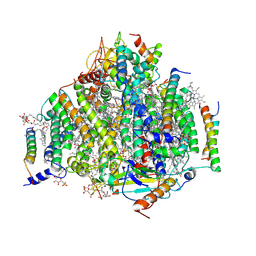 | | PSII intermediate Psb28-RC47 | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Sui, S.F, Shen, J.R, Han, G.Y, Xiao, Y.N, Huang, G.Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into cyanobacterial photosystem II intermediates associated with Psb28 and Tsl0063.
Nat.Plants, 7, 2021
|
|
7DTC
 
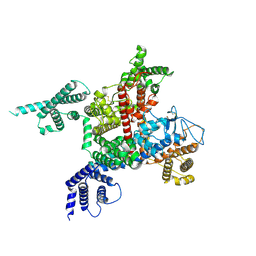 | | voltage-gated sodium channel Nav1.5-E1784K | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Pan, X, Li, Z. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na v 1.5 reveals the fast inactivation-related segments as a mutational hotspot for the long QT syndrome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DNT
 
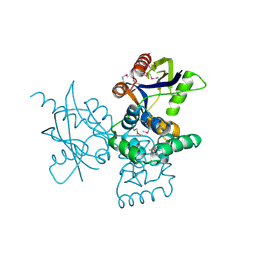 | | mRNA-decapping enzyme g5Rp | | Descriptor: | mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2022-03-09 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7CZL
 
 | | Structural insights into a dimeric Psb27-photosystem II complex from a cyanobacterium Thermosynechococcus vulcanus | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pi, X, Huang, G, Xiao, Y. | | Deposit date: | 2020-09-09 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural insights into a dimeric Psb27-photosystem II complex from a cyanobacterium Thermosynechococcus vulcanus .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5U6G
 
 | |
