5DFM
 
 | | Structure of Tetrahymena telomerase p19 fused to MBP | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Telomerase-associated protein 19, SULFATE ION, ... | | Authors: | Chan, H, Cascio, D, Sawaya, M.R, Feigon, J. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions.
Science, 350, 2015
|
|
1HKO
 
 | | NMR structure of bovine cytochrome b5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muskett, F.W, Whitford, D. | | Deposit date: | 2003-03-10 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bovine Ferricytochrome B5 Determined Using Heteronuclear NMR Methods.
J.Mol.Biol., 258, 1996
|
|
1HTD
 
 | | STRUCTURAL INTERACTION OF NATURAL AND SYNTHETIC INHIBITORS WITH THE VENOM METALLOPROTEINASE, ATROLYSIN C (HT-D) | | Descriptor: | ATROLYSIN C, CALCIUM ION, ZINC ION | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1994-01-20 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1IRX
 
 | | Crystal structure of class I lysyl-tRNA synthetase | | Descriptor: | ZINC ION, lysyl-tRNA synthetase | | Authors: | Nureki, O, Terada, T, Ishitani, R, Ambrogelly, A, Ibba, M, Soll, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-10-25 | | Release date: | 2002-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional convergence of two lysyl-tRNA synthetases with unrelated topologies.
Nat.Struct.Biol., 9, 2002
|
|
1IV6
 
 | | Solution Structure of the DNA Complex of Human TRF1 | | Descriptor: | 5'-D(*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3', TELOMERIC REPEAT BINDING FACTOR 1 | | Authors: | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a telomeric DNA complex of human TRF1.
Structure, 9, 2001
|
|
4JC3
 
 | |
1J8Z
 
 | | Solution structure of beta3 analogue peptide (HCYS) of HIV gp41 600-612 loop. | | Descriptor: | HCYS BETA3-CYS ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-05-23 | | Release date: | 2003-07-01 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1LFW
 
 | | Crystal structure of pepV | | Descriptor: | 3-[(1-AMINO-2-CARBOXY-ETHYL)-HYDROXY-PHOSPHINOYL]-2-METHYL-PROPIONIC ACID, ZINC ION, pepV | | Authors: | Jozic, D, Bourenkow, G, Bartunik, H, Scholze, H, Dive, V, Henrich, B, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Dinuclear Zinc Aminopeptidase PepV from Lactobacillus delbrueckii Unravels Its Preference for Dipeptides
Structure, 10, 2002
|
|
1LLW
 
 | | Structural studies on the synchronization of catalytic centers in glutamate synthase: complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | van den Heuvel, R.H, Ferrari, D, Bossi, R.T, Ravasio, S, Curti, B, Vanoni, M.A, Florencio, F.J, Mattevi, A. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on the synchronization of catalytic centers in glutamate synthase
J.BIOL.CHEM., 277, 2002
|
|
1LS8
 
 | | NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH | | Descriptor: | pheromone binding protein | | Authors: | Lee, D, Damberger, F, Horst, R, Guntert, P, Leal, W.S, Wuthrich, K. | | Deposit date: | 2002-05-17 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH
FEBS Lett., 531, 2002
|
|
1LU4
 
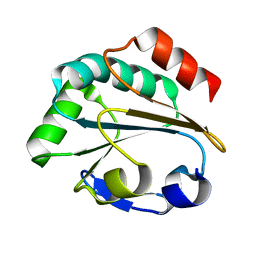 | | 1.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED MYCOBACTERIUM TUBERCULOSIS DISULFIDE OXIDOREDUCTASE HOMOLOGOUS TO E. COLI DSBE: IMPLICATIONS FOR FUNCTIONS | | Descriptor: | SOLUBLE SECRETED ANTIGEN MPT53 | | Authors: | Goulding, C.W, Apostol, M.I, Gleiter, S, Parseghian, A, Bardwell, J, Gennaro, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-21 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Gram-positive DsbE Proteins Function Differently from Gram-negative DsbE Homologs: A STRUCTURE TO FUNCTION ANALYSIS OF DsbE FROM MYCOBACTERIUM TUBERCULOSIS.
J.Biol.Chem., 279, 2004
|
|
1L3C
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP WITH SHORT CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of Mt0146/Cbit Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L6V
 
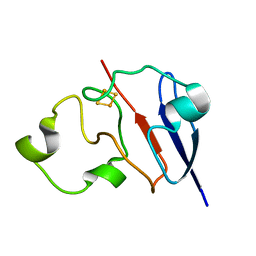 | | STRUCTURE OF REDUCED BOVINE ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
1L7L
 
 | | Crystal structure of Pseudomonas aeruginosa lectin 1 determined by single wavelength anomalous scattering phasing method | | Descriptor: | CALCIUM ION, PA-I galactophilic lectin | | Authors: | Liu, Z.J, Tempel, W, Lin, D, Karaveg, K, Doyle, R.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-03-15 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure determination of P. aeruginosa lectin-1 using single
wavelength anomalous scattering data from native crystals (P028)
AM.CRYST.ASSOC.,ABSTR.PAPERS (ANNUAL MEETING), 29, 2002
|
|
1L8I
 
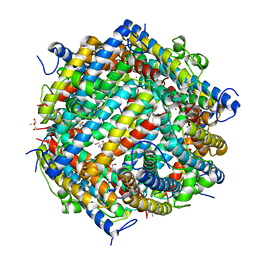 | | Dna Protection and Binding by E. Coli DPS Protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, POTASSIUM ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2002-03-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA Protection and Binding by E. Coli Dps Protein
To be Published
|
|
1L4V
 
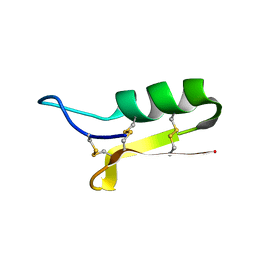 | | SOLUTION STRUCTURE OF SAPECIN | | Descriptor: | Sapecin | | Authors: | Hanzawa, H, Iwai, H, Takeuchi, K, Kuzuhara, T, Komano, H, Kohda, D, Inagaki, F, Natori, S, Arata, Y, Shimada, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H nuclear magnetic resonance study of the solution conformation of an antibacterial protein, sapecin.
FEBS Lett., 269, 1990
|
|
1L5X
 
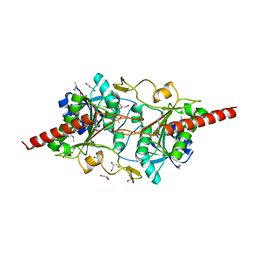 | | The 2.0-Angstrom resolution crystal structure of a survival protein E (SurE) homolog from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, Survival protein E | | Authors: | Mura, C, Katz, J.E, Clarke, S.G, Eisenberg, D. | | Deposit date: | 2002-03-08 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of an Archaeal Homolog of Survival
Protein E (SurE-alpha): An Acid Phosphatase with Purine
Nucleotide Specificity
J.Mol.Biol., 326, 2003
|
|
4HFR
 
 | | Human 11beta-Hydroxysteroid Dehydrogenase Type 1 in complex with an orally bioavailable acidic inhibitor AZD4017. | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {(3S)-1-[5-(cyclohexylcarbamoyl)-6-(propylsulfanyl)pyridin-2-yl]piperidin-3-yl}acetic acid | | Authors: | Ogg, D.J, Gerhardt, S, Hargreaves, D. | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Bioavailable Acidic 11 -Hydroxysteroid Dehydrogenase Type 1 (11 -HSD1) Inhibitor: Discovery of 2-[(3S)-1-[5-(Cyclohexylcarbamoyl)-6-propylsulfanylpyridin-2-yl]-3-piperidyl]acetic Acid (AZD4017)
J.Med.Chem., 55, 2012
|
|
1IIZ
 
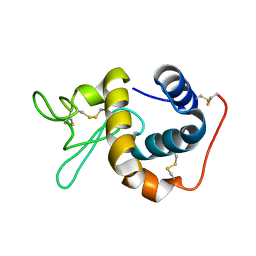 | | Crystal Structure of the Induced Antibacterial Protein from Tasar Silkworm, Antheraea mylitta | | Descriptor: | LYSOZYME | | Authors: | Jain, D, Nair, D.T, Swaminathan, G.J, Abraham, E.G, Nagaraju, J, Salunke, D.M. | | Deposit date: | 2001-04-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the induced antibacterial protein from tasar silkworm, Antheraea mylitta. Implications to molecular evolution.
J.Biol.Chem., 276, 2001
|
|
1IY1
 
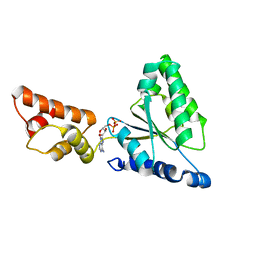 | | Crystal structure of the FtsH ATPase domain with ADP from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent metalloprotease FtsH | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
4N6C
 
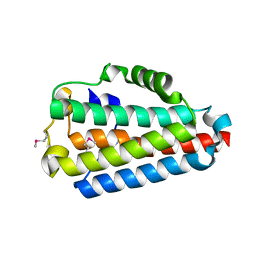 | | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium (NESG) Target SpR36. | | Descriptor: | BROMIDE ION, uncharacterized protein | | Authors: | Vorobiev, S, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae.
To be Published
|
|
1IE8
 
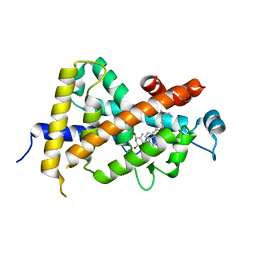 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IEH
 
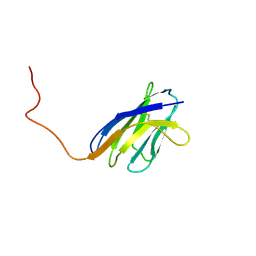 | | SOLUTION STRUCTURE OF A SOLUBLE SINGLE-DOMAIN ANTIBODY WITH HYDROPHOBIC RESIDUES TYPICAL OF A VL/VH INTERFACE | | Descriptor: | BRUC.D4.4 | | Authors: | Vranken, W, Tolkatchev, D, Xu, P, Tanha, J, Chen, Z, Narang, S, Ni, F. | | Deposit date: | 2001-04-09 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a llama single-domain antibody with hydrophobic residues typical of the VH/VL interface.
Biochemistry, 41, 2002
|
|
1L0L
 
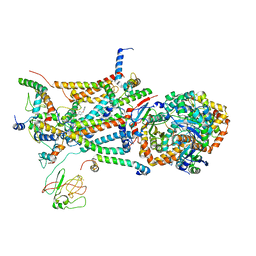 | | structure of bovine mitochondrial cytochrome bc1 complex with a bound fungicide famoxadone | | Descriptor: | Cytochrome B, Cytochrome c1, heme protein, ... | | Authors: | Gao, X, Wen, X, Yu, C.A, Esser, L, Tsao, S, Quinn, B, Zhang, L, Yu, L, Xia, D. | | Deposit date: | 2002-02-11 | | Release date: | 2003-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of Mitochondrial Cytochrome bc1 in Complex with Famoxadone: The Role of Aromatic-Aromatic Interaction in Inhibition
Biochemistry, 41, 2003
|
|
1L3B
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP W/ LONG CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
