7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
3R1G
 
 | | Structure Basis of Allosteric Inhibition of BACE1 by an Exosite-Binding Antibody | | Descriptor: | Beta-secretase 1, FAB of YW412.8.31 antibody heavy chain, FAB of YW412.8.31 antibody light chain | | Authors: | Wang, W, Rouge, L, Wu, P, Chiu, C, Chen, Y, Wu, Y, Watts, R.J. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-{beta} Production in Vivo.
Sci Transl Med, 3, 2011
|
|
5YN8
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNJ
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNI
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNF
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
6JJ6
 
 | | BRD4 in complex with 500 | | Descriptor: | 1-methyl-6-(3-(4-methylpiperazine-1-carbonyl)benzyl)-1,2a1,5a,6-tetrahydro-2H-pyrido[3',2':6,7]azepino[4,3,2-cd]isoindol-2-one, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | BRD4 in complex with ZZM1
To Be Published
|
|
5YNB
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to Sinefungin | | Descriptor: | SINEFUNGIN, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
6JBJ
 
 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | Authors: | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
6JJ5
 
 | | BRD4 in complex with 259 | | Descriptor: | 3-((1-methyl-2-oxo-1,2,2a1,5a-tetrahydro-6H-pyrido[3',2':6,7]azepino[4,3,2-cd]isoindol-6-yl)methyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | BRD4 in complex with ZZM1
To Be Published
|
|
5Z74
 
 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alr0819 protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
5Z9J
 
 | | Identification of the functions of unusual cytochrome p450-like monooxygenases involved in microbial secondary metablism | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like enzyme, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lu, M, Lin, L, Zhang, C, Chen, Y. | | Deposit date: | 2018-02-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Riboflavin Is Directly Involved in the N-Dealkylation Catalyzed by Bacterial Cytochrome P450 Monooxygenases.
Chembiochem, 2020
|
|
8YDD
 
 | | Crystal structure of Nanog(R100A) in complex with Wdr5 | | Descriptor: | Homeobox protein NANOG, WD repeat-containing protein 5 | | Authors: | Wang, D, Shi, X, Zhao, L, Wu, B, Chen, Y. | | Deposit date: | 2024-02-20 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Nanog(R100A) in complex with Wdr5
To Be Published
|
|
6ACS
 
 | | poly-cis-prenyltransferase | | Descriptor: | CITRIC ACID, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific), GLYCEROL, ... | | Authors: | Ko, T.-P, Chen, Y. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of undecaprenyl pyrophosphate synthase from Acinetobacter baumannii
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5Z73
 
 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 | | Descriptor: | Alr0819 protein, GLYCEROL | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
5Z9I
 
 | |
5ZLT
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Ko, T.P, Hsieh, T.J, Yang, C.S, Chen, Y. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
7EBL
 
 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), STING | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
7EBD
 
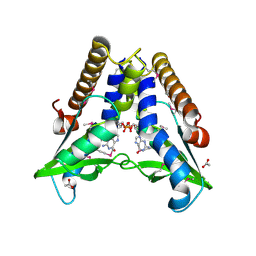 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, TIR-like domain-containing protein | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
6J3Q
 
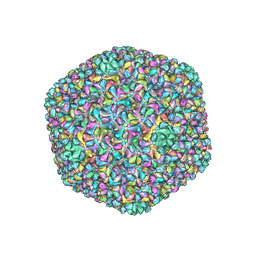 | | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | | Descriptor: | cement protein, major capsid protein | | Authors: | Jin, H, Jiang, Y.L, Yang, F, Zhang, J.T, Li, W.F, Zhou, K, Ju, J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-05 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1.
Structure, 27, 2019
|
|
6KKM
 
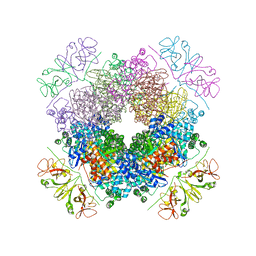 | | Crystal structure of RbcL-Raf1 complex from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein, Ribulose bisphosphate carboxylase large chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-07-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
7XBK
 
 | | Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform 2 of Caseinolytic peptidase B protein homolog, MAGNESIUM ION, ... | | Authors: | Wu, D, Liu, Y, Dai, Y, Wang, G, Lu, G, Chen, Y, Li, N, Lin, J, Gao, N. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
6KKN
 
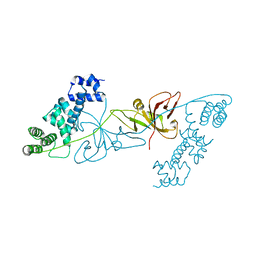 | | Crystal structure of RuBisCO accumulation factor Raf1 from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-07-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
7X3O
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054 | | Descriptor: | (2~{R})-2-(3-fluoranyl-4-pyrimidin-5-yl-phenyl)butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054
To Be Published
|
|
