8G93
 
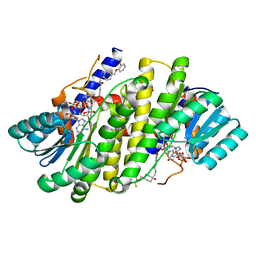 | | Crystal structures of 17-beta-hydroxysteroid dehydrogenase 13 | | Descriptor: | 3-fluoro-N-({(1r,4r)-4-[(2-fluorophenoxy)methyl]-1-hydroxycyclohexyl}methyl)-4-hydroxybenzamide, Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G84
 
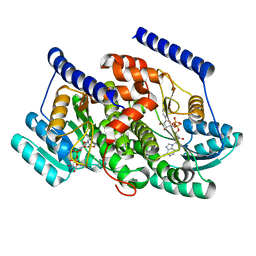 | | Crystal structures of HSD17B13 complexes | | Descriptor: | Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G89
 
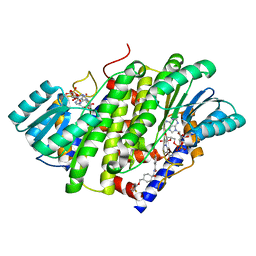 | | HSD17B13 in complex with cofactor and inhibitor | | Descriptor: | 3-fluoro-N-({(1r,4r)-4-[(2-fluorophenoxy)methyl]-1-hydroxycyclohexyl}methyl)-4-hydroxybenzamide, Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
4YZK
 
 | |
8QQI
 
 | |
4YZL
 
 | | Crystal structure of the indole prenyltransferase TleC complexed with indolactam V and DMSPP | | Descriptor: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Tryptophan dimethylallyltransferase | | Authors: | Mori, T, Morita, H, Abe, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
4PSW
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
1ULW
 
 | | Crystal structure of P450nor Ser73Gly/Ser75Gly mutant | | Descriptor: | Cytochrome P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Su, F, Li, Z, Takaya, N, Shoun, H. | | Deposit date: | 2003-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
4AQ3
 
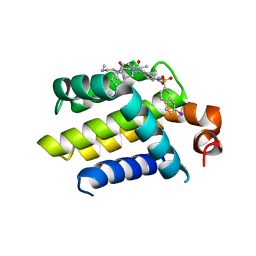 | | HUMAN BCL-2 WITH PHENYLACYLSULFONAMIDE INHIBITOR | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, N,N-dibutyl-4-chloranyl-1-[2-(3,4-dihydro-1H-isoquinolin-2-ylcarbonyl)-4-[(7-iodanylnaphthalen-2-yl)sulfonylcarbamoyl]phenyl]-5-methyl-pyrazole-3-carboxamide | | Authors: | Bertrand, J.A, Fasolini, M, Modugno, M. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a Phenylacylsulfonamide Series of Dual Bcl-2/Bcl-Xl Antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4YZJ
 
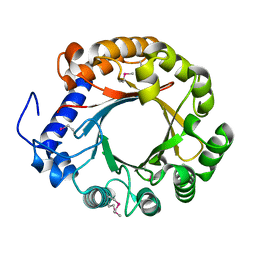 | |
3ROP
 
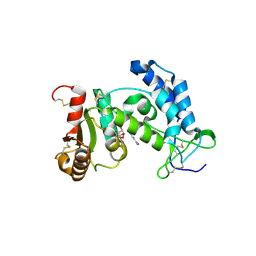 | |
3ROK
 
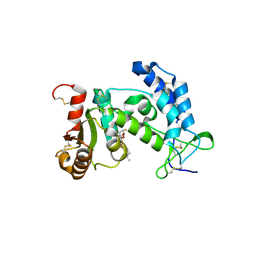 | |
4FZD
 
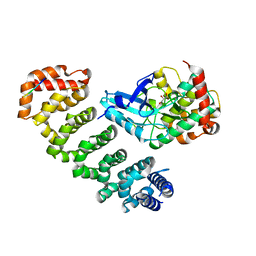 | | Crystal structure of MST4-MO25 complex with WSF motif | | Descriptor: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
4Q9V
 
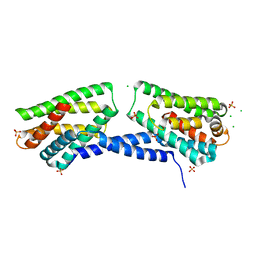 | | Crystal structure of TIPE3 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | Authors: | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
6UMJ
 
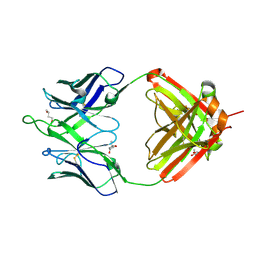 | | Crystal structure of erenumab Fab-c | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-BUTANEDIOL, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMG
 
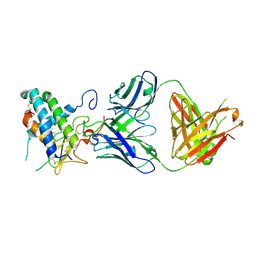 | | Crystal structure of erenumab Fab bound to the extracellular domain of CGRP receptor | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 1, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
7E24
 
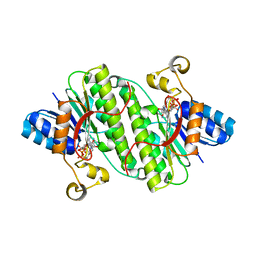 | |
7E28
 
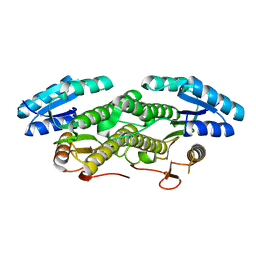 | |
8UNH
 
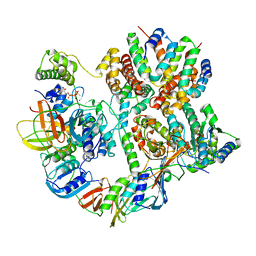 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-19 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
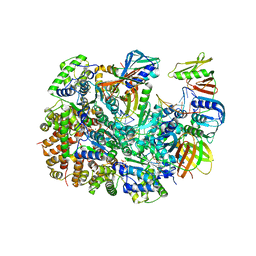 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J0P
 
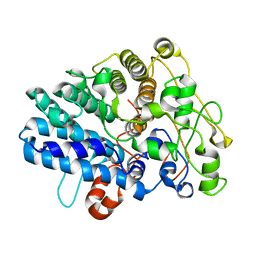 | | Chitin binding SusD-like protein AqSusD from a marine Bacteroidetes | | Descriptor: | Chitin binding SusD-like protein | | Authors: | Yang, J. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
8JNC
 
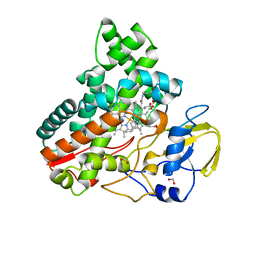 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-maltophilin | | Descriptor: | (1Z,3E,5S,8R,9S,10S,11R,13R,15R,16S,18Z,24S,25S)-11-ethyl-2,24-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-7,20,27,28-tetraone, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNQ
 
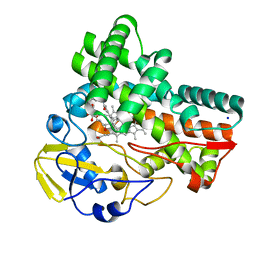 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with a substrate compound c | | Descriptor: | (1Z,3E,5E,7S,8R,10S,11R,13R,15R,16E,18E,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.09,13.08,15]octacosa-1(2),3,5,16,18-pentaene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
