6CFD
 
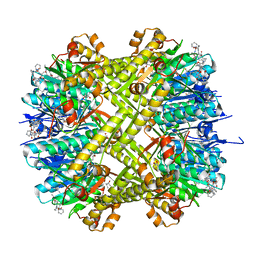 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
5Z6Z
 
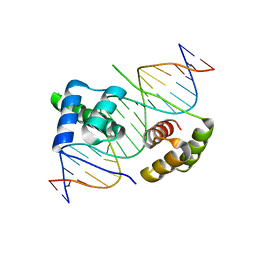 | | Crystal structure of human DUX4 homeodomains bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-01-26 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
6BMH
 
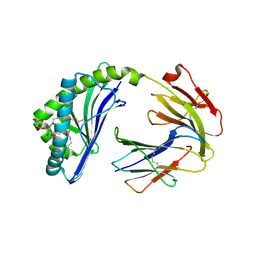 | | Crystal structure of MHC-I like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d2, ... | | Authors: | Khandokar, Y.B, Le Nours, J, Rossjohn, J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Differing roles of CD1d2 and CD1d1 proteins in type I natural killer T cell development and function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BMK
 
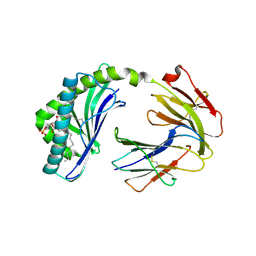 | | Crystal structure of MHC-I like protein | | Descriptor: | (2R)-1-(decanoyloxy)-3-(phosphonooxy)propan-2-yl octadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Khandokar, Y.B, Le Nours, J, Rossjohn, J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Differing roles of CD1d2 and CD1d1 proteins in type I natural killer T cell development and function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3ZXH
 
 | |
5Y4T
 
 | | Crystal structure of Trx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutaredoxin | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
5Y9C
 
 | |
8HAW
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
5Y9E
 
 | | Crystal structure of HPV58 pentamer | | Descriptor: | GLYCEROL, MAGNESIUM ION, Major capsid protein L1 | | Authors: | Li, S.W, Li, Z.H. | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Crystal Structures of Two Immune Complexes Identify Determinants for Viral Infectivity and Type-Specific Neutralization of Human Papillomavirus.
MBio, 8, 2017
|
|
8HAV
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8H3X
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 282 | | Descriptor: | Fragilysin, ZINC ION, nanobody 282 | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8H3Y
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 327 | | Descriptor: | Fragilysin, Nanobody 327, ZINC ION | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
7YW0
 
 | | Bacteroides fragilis Hcp5 | | Descriptor: | Bacterodales T6SS protein TssD (Hcp) | | Authors: | Wen, Y, He, W, Bai, Y. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and assembly of type VI secretion system cargo delivery vehicle.
Cell Rep, 42, 2023
|
|
8HQ8
 
 | | Bry-LHCII homotrimer of Bryopsis corticulans | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2022-12-13 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional properties of different types of siphonous LHCII trimers from an intertidal green alga Bryopsis corticulans.
Structure, 31, 2023
|
|
8HPD
 
 | | Bry-LHCII heterotrimer of Bryopsis corticulans | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional properties of different types of siphonous LHCII trimers from an intertidal green alga Bryopsis corticulans.
Structure, 31, 2023
|
|
8HLV
 
 | | Bry-LHCII homotrimer of Bryopsis corticulans | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural and functional properties of different types of siphonous LHCII trimers from an intertidal green alga Bryopsis corticulans.
Structure, 31, 2023
|
|
2Z8O
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, L(+)-TARTARIC ACID | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
8ECG
 
 | | Complex of HMG1 with pitavastatin | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1, GLYCEROL, Pitavastatin | | Authors: | Haywood, J, Bond, C.S. | | Deposit date: | 2022-09-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.
Nat Commun, 13, 2022
|
|
2Z8P
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | (GLY)(GLU)(ALA)(TPO)(VAL)(PTR)(ALA), 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5Y9F
 
 | |
5Y4U
 
 | | Crystal structure of Grx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | Monothiol glutaredoxin-3 | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
1ROW
 
 | | Structure of SSP-19, an MSP-domain protein like family member in Caenorhabditis elegans | | Descriptor: | MSP-domain protein like family member | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, J, Lu, S, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sperm-specific protein SSP-19 from Caenorhabditis elegans.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2Z3C
 
 | |
2Z3D
 
 | |
