1ZBY
 
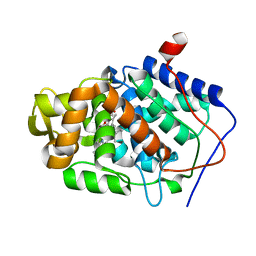 | | High-Resolution Crystal Structure of Native (Resting) Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Bhaskar, B, Shimizu, H, Li, H, Sundaramoorthy, M, McRee, D.E, Goodin, D.B, Poulos, T.L. | | Deposit date: | 2005-04-09 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase
Biochemistry, 42, 2003
|
|
1AP8
 
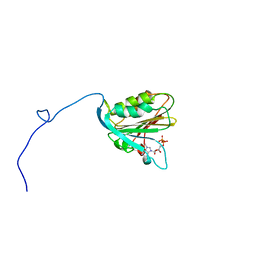 | | TRANSLATION INITIATION FACTOR EIF4E IN COMPLEX WITH M7GDP, NMR, 20 STRUCTURES | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR EIF4E | | Authors: | Matsuo, H, Li, H, Mcguire, A.M, Fletcher, M, Gingras, A.C, Sonenberg, N, Wagner, G. | | Deposit date: | 1997-07-25 | | Release date: | 1998-01-28 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein.
Nat.Struct.Biol., 4, 1997
|
|
1ZBZ
 
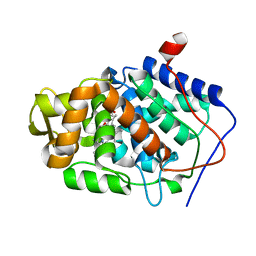 | | High-Resolution Crystal Structure of Compound I intermediate of Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Bhaskar, B, Shimizu, H, Li, H, Sundaramoorthy, M, McRee, D.E, Goodin, D.B, Poulos, T.L. | | Deposit date: | 2005-04-09 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase
Biochemistry, 42, 2003
|
|
3KRD
 
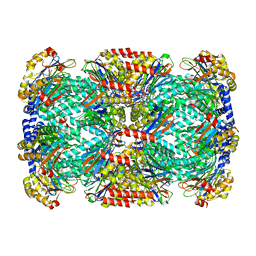 | |
5THO
 
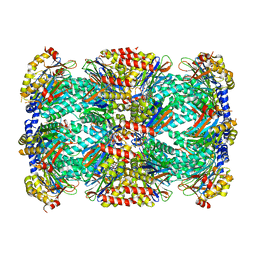 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome in complex with N,C-capped Dipeptide Inhibitor PKS2205 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2016-09-30 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
6PP4
 
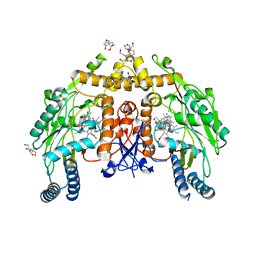 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(pyridin-3-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(pyridin-3-yl)methoxy]phenyl}-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
7TFL
 
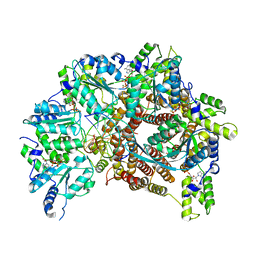 | | Atomic model of S. cerevisiae clamp loader RFC bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFI
 
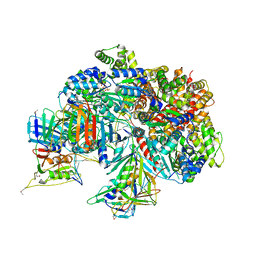 | | Atomic model of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with an open clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFH
 
 | | Atomic model of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFJ
 
 | | Atomic model of S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with a closed clamp ring | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFK
 
 | | Atomic model of S. cerevisiae clamp loader RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
6P2R
 
 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6P28
 
 | |
6P25
 
 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6PP2
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 4-((4-(2-Amino-4-methylquinolin-7-yl)-2-(aminomethyl)phenoxy)methyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{[2-(aminomethyl)-4-(2-amino-4-methylquinolin-7-yl)phenoxy]methyl}benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6POW
 
 | | Structure of human endotheial nitric oxide synthase heme domain in complex with 7-(5-(Aminomethyl)pyridin-3-yl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[5-(aminomethyl)pyridin-3-yl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6POZ
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-isopropoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(propan-2-yl)oxy]phenyl}-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6POX
 
 | | Structure of human endothelialnitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-ethoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-ethoxyphenyl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6POY
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-propoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-propoxyphenyl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6POV
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(2-Aminoethyl)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)phenyl]-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
8K5R
 
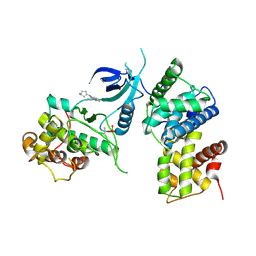 | | CDK9/cyclin T1 in complex with KB-0742 | | Descriptor: | (1S,3S)-N3-(5-pentan-3-ylpyrazolo[1,5-a]pyrimidin-7-yl)cyclopentane-1,3-diamine, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Zhou, M, Li, H, Gao, H, Trotter, B.W, Freeman, D. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Discovery of KB-0742, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of CDK9 for MYC-Dependent Cancers.
J.Med.Chem., 66, 2023
|
|
7LHG
 
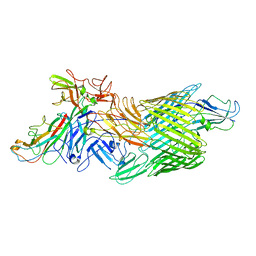 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapG in the first conformation | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, P fimbria tip G-adhesin PapG-II, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
7LHH
 
 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapG in the second conformation | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, P fimbria tip G-adhesin PapG-II, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
7LHI
 
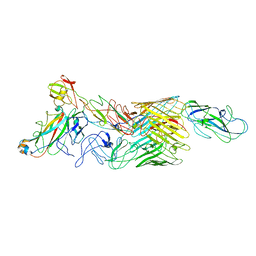 | | Cryo-EM structure of E. coli P pilus tip assembly intermediate PapC-PapD-PapK-PapF-PapG | | Descriptor: | Chaperone protein PapD, Fimbrial adapter PapK, Fimbrial protein, ... | | Authors: | Du, M, Yuan, Z, Werneburg, G, Henderson, N, Chauhan, H, Kovach, A, Zhao, G, Johl, J, Li, H, Thanassi, D. | | Deposit date: | 2021-01-23 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis.
Nat Commun, 12, 2021
|
|
6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
