8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
7CX4
 
 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX3
 
 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX2
 
 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
4L6X
 
 | |
4L0G
 
 | | Crystal Structure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside hydrolase family 48, ... | | Authors: | Jiao, A, Bai, A, Yan, F, Geng, W. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary X-ray analysis of a processive cellobiohydrolase CbCBH48A from Caldicellulosiruptor bescii
to be published
|
|
1ZZ6
 
 | | Crystal Structure of Apo-HppE | | Descriptor: | Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZB
 
 | | Crystal Structure of CoII HppE in Complex with Substrate | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, COBALT (II) ION, Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ8
 
 | | Crystal Structure of FeII HppE in Complex with Substrate Form 2 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZC
 
 | | Crystal Structure of CoII HppE in Complex with Tris Buffer | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT (II) ION, hydroxypropylphosphonic acid epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ7
 
 | | Crystal Structure of FeII HppE in Complex with Substrate form 1 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxyprophylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ9
 
 | | Crystal Structure of FeII HppE | | Descriptor: | FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase, SULFATE ION | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1KXT
 
 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
7F4A
 
 | |
7F3S
 
 | |
7F51
 
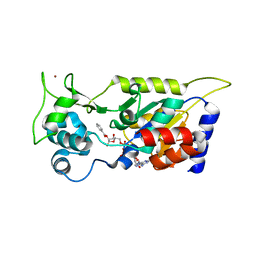 | | Crystal structure of Hst2 in complex with 2'-O-Benzoyl ADP Ribose | | Descriptor: | NAD-dependent protein deacetylase HST2, ZINC ION, [(2R,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] benzoate | | Authors: | Wang, D, Yan, F, Chen, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Global profiling of regulatory elements in the histone benzoylation pathway.
Nat Commun, 13, 2022
|
|
7F4E
 
 | |
7F5M
 
 | |
1CZ9
 
 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN (D64N) FROM CITRATE | | Descriptor: | AVIAN SARCOMA VIRUS INTEGRASE, CITRIC ACID, SULFATE ION | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-09-01 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
5J8R
 
 | | Crystal Structure of the Catalytic Domain of Human Protein Tyrosine Phosphatase non-receptor Type 12 - K61R mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Li, H, Yang, F, Xu, Y.F, Wang, W.J, Xiao, P, Yu, X, Sun, J.P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Crystal structure and substrate specificity of PTPN12.
Cell Rep, 15, 2016
|
|
1CXU
 
 | | 1.42A RESOLUTION ASV INTEGRASE CORE DOMAIN FROM CITRATE | | Descriptor: | CITRIC ACID, GLYCEROL, PROTEIN (AVIAN SARCOMA VIRUS INTEGRASE) | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1999-08-30 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
