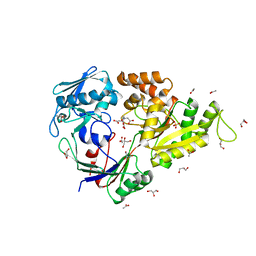
5L8D
 
 | |
6DFG
 
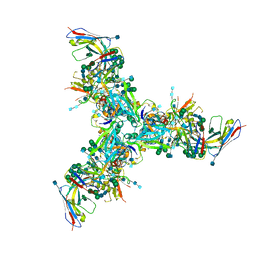 | | BG505 MD39 SOSIP trimer in complex with mature BG18 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Steichen, J.M, Schief, W.R, Ward, A.B. | | Deposit date: | 2018-05-14 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
7NAY
 
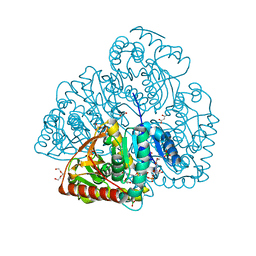 | | Crystal structure of lactate dehydrogenase from Selenomonas ruminantium with NADH. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, L-lactate dehydrogenase, ... | | Authors: | Bertrand, Q, Robin, A, Girard, E, Madern, D. | | Deposit date: | 2021-01-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
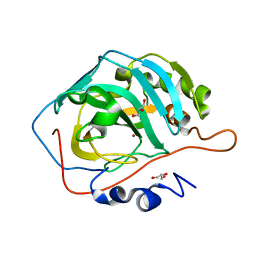
3U3A
 
 | |
5LBT
 
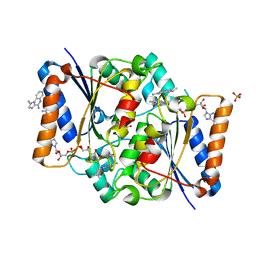 | | Structure of the human quinone reductase 2 (NQO2) in complex with imiquimod | | Descriptor: | 1-(2-methylpropyl)imidazo[4,5-c]quinolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Gross, O. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Imiquimod Inhibits Mitochondrial Complex I and Induces K+ efflux-independent Nlrp3 Inflammasome Activation via Nek7
To Be Published
|
|
5LBU
 
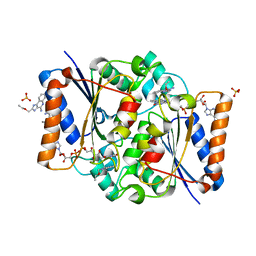 | | Structure of the human quinone reductase 2 (NQO2) in complex with to CL097 | | Descriptor: | 2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Gross, O. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imiquimod Inhibits Mitochondrial Complex I and Induces K+ efflux-independent Nlrp3 Inflammasome Activation via Nek7
To Be Published
|
|
5DZA
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA C terminal domain (WT) | | Descriptor: | 1,2-ETHANEDIOL, BspA, DI(HYDROXYETHYL)ETHER | | Authors: | Rego, S, Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
5DZ9
 
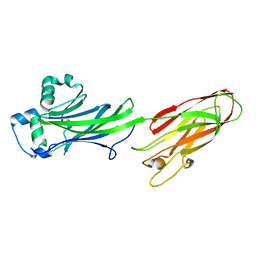 | |
4FCB
 
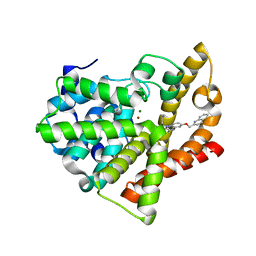 | | Potent and Selective Phosphodiesterase 10A Inhibitors | | Descriptor: | 3,4-dimethyl-1-propyl-7-(quinolin-2-ylmethoxy)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazines as potent and selective phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4FCD
 
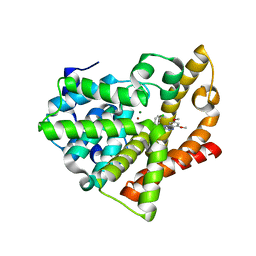 | | Potent and Selective Phosphodiesterase 10A Inhibitors | | Descriptor: | 1-(2-chlorophenyl)-6,8-dimethoxy-3-methylimidazo[5,1-c][1,2,4]benzotriazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Novel triazines as potent and selective phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5JP2
 
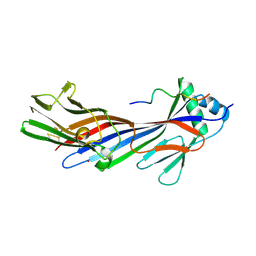 | |
9FH9
 
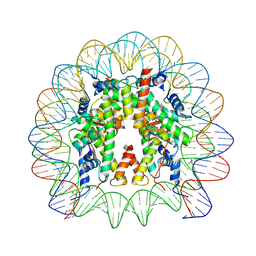 | |
4UPH
 
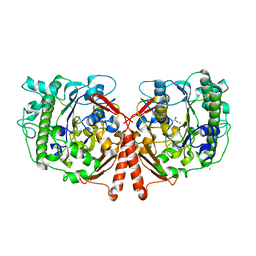 | | Crystal Structure of Phosphonate Monoester Hydrolase of Agrobacterium radiobacter | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATASE (SULFURIC ESTER HYDROLASE) PROTEIN | | Authors: | Fischer, G, Loo, B.v, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
4UPL
 
 | | Dimeric sulfatase SpAS2 from Silicibacter pomeroyi | | Descriptor: | SULFATASE FAMILY PROTEIN, ZINC ION | | Authors: | Jonas, S, van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
4UPI
 
 | | Dimeric sulfatase SpAS1 from Silicibacter pomeroyi | | Descriptor: | SULFATASE FAMILY PROTEIN, ZINC ION | | Authors: | Jonas, S, van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
4UPK
 
 | |
6YEK
 
 | | Crystal structure of human NEMO apo form | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b | | Authors: | Garcia-Pardo, J, Akutsu, M, Busse, P, Skenderovic, A, Maculins, T, Dikic, I. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
6XX0
 
 | | Crystal structure of NEMO in complex with Ubv-LIN | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b, ... | | Authors: | Akutsu, M, Skenderovic, A, Garcia-Pardo, J, Maculins, T, Dikic, I. | | Deposit date: | 2020-01-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
9FGQ
 
 | |
3V6E
 
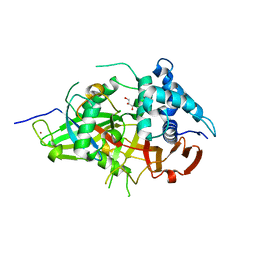 | | Crystal Structure of USP2 and a mutant form of Ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
6NF5
 
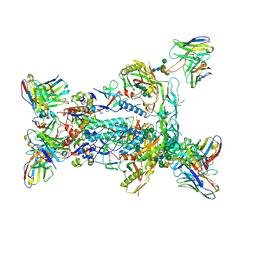 | | BG505 MD64 N332-GT5 SOSIP trimer in complex with BG18-like precursor HMP1 fragmentantigen binding and base-binding RM20A3 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG18-like precursor HMP1 fragment antigen binding heavy chain, ... | | Authors: | Ozorowski, G, Torres, J.L, Steichen, J.M, Schief, W.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
6NFC
 
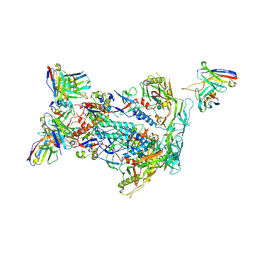 | | BG505 MD64 N332-GT5 SOSIP trimer in complex with BG18-like precursor HMP42 fragmentantigen binding and base-binding RM20A3 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG18-like precursor HMP42 fragment antigen binding heavy chain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-19 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
5M4N
 
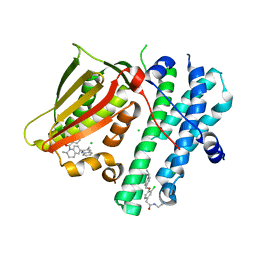 | | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase | | Descriptor: | CHLORIDE ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [2,4-bis(oxidanyl)phenyl]-[(2~{S})-2-methyl-6-(3-methylquinolin-2-yl)-3,4-dihydro-2~{H}-quinolin-1-yl]methanone, ... | | Authors: | Baker, L.M, Brough, P, Surgenor, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase.
J. Med. Chem., 60, 2017
|
|
5M4P
 
 | | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase | | Descriptor: | CHLORIDE ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, ... | | Authors: | Baker, L.M, Brough, P, Surgenor, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase.
J. Med. Chem., 60, 2017
|
|
5M4K
 
 | | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, ... | | Authors: | Baker, L.M, Brough, P, Surgenor, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase.
J. Med. Chem., 60, 2017
|
|
