1R2S
 
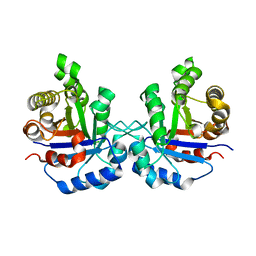 | |
1R8O
 
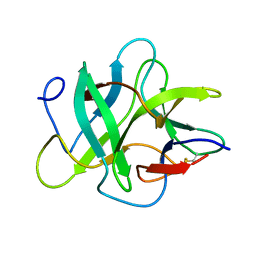 | | Crystal structure of an unusual Kunitz-type trypsin inhibitor from Copaifera langsdorffii seeds | | Descriptor: | Kunitz trypsin inhibitor | | Authors: | Krauchenco, S, Nagem, R.A.P, da Silva, J.A, Marangoni, S, Polikarpov, I. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Three-dimensional structure of an unusual Kunitz (STI) type trypsin inhibitor from Copaifera langsdorffii.
Biochimie, 86, 2004
|
|
1R2T
 
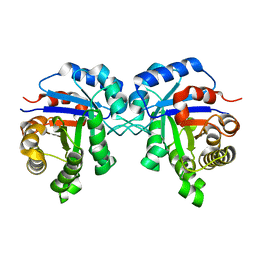 | |
1R2R
 
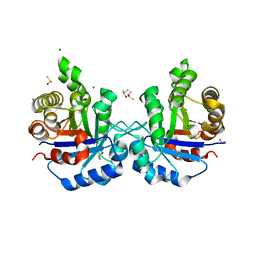 | | CRYSTAL STRUCTURE OF RABBIT MUSCLE TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Aparicio, R, Ferreira, S.T, Polikarpov, I. | | Deposit date: | 2003-09-29 | | Release date: | 2003-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Closed conformation of the active site loop of rabbit muscle triosephosphate isomerase in the absence of substrate: evidence of conformational heterogeneity.
J.Mol.Biol., 334, 2003
|
|
1GMZ
 
 | |
1GQN
 
 | | Native 3-dehydroquinase from Salmonella typhi | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Lee, W.-H, Perles, L.A, Nagem, R.A.P, Polikarpov, I, Sawyer, L. | | Deposit date: | 2001-11-26 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Comparison of Different Crystal Forms of 3-Dehydroquinase from Salmonella Typhi and its Implications for Enzyme Activity
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HWU
 
 | | STRUCTURE OF PII PROTEIN FROM HERBASPIRILLUM SEROPEDICAE | | Descriptor: | PII PROTEIN | | Authors: | Benelli, E.M, Buck, M, Polikarpov, I, De Souza, E.M, Cruz, L.M, Pedrosa, F.O. | | Deposit date: | 2001-01-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Herbaspirillum seropedicae signal transduction protein PII is structurally similar to the enteric GlnK.
Eur.J.Biochem., 269, 2002
|
|
2H79
 
 | | Crystal Structure of human TR alpha bound T3 in orthorhombic space group | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, THRA protein | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Bleicher, L, Ambrosio, A.L.B, Figueira, A.C.M, Santos, M.A.M, Neto, M.O, Fischer, H, Togashi, H.F.M, Craievich, A.F, Garrat, R.C, Baxter, J.D, Webb, P, Polikarpov, I. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function.
J.Mol.Biol., 360, 2006
|
|
2H77
 
 | | Crystal structure of human TR alpha bound T3 in monoclinic space group | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, THRA protein | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Bleicher, L, Ambrosio, A.L.B, Figueira, A.C.M, Santos, M.A.M, Neto, M.O, Fischer, H, Togashi, H.F.M, Craievich, A.F, Garrat, R.C, Baxter, J.D, Webb, P, Polikarpov, I. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function.
J.Mol.Biol., 360, 2006
|
|
2H9T
 
 | | Crystal structure of human alpha-thrombin in complex with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, PPACK active site thrombin inhibitor, Thrombin | | Authors: | Lima, L.M.T.R, Polikarpov, I, Monteiro, R.Q. | | Deposit date: | 2006-06-11 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and thermodynamic analysis of thrombin:suramin interaction in solution and crystal phases.
Biochim.Biophys.Acta, 1794, 2009
|
|
1L9W
 
 | | CRYSTAL STRUCTURE OF 3-DEHYDROQUINASE FROM SALMONELLA TYPHI COMPLEXED WITH REACTION PRODUCT | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase aroD | | Authors: | Lee, W.H, Perles, L.A, Nagem, R.A.P, Shrive, A.K, Hawkins, A, Sawyer, L, Polikarpov, I. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of different crystal forms of 3-dehydroquinase from Salmonella typhi and its implication for the enzyme activity.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3SZ1
 
 | | Human PPAR gamma ligand binding domain in complex with luteolin and myristic acid | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, MYRISTIC ACID, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Puhl, A.C, Bernardes, A, Polikarpov, I. | | Deposit date: | 2011-07-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mode of peroxisome proliferator-activated receptor gamma activation by luteolin.
Mol.Pharmacol., 81, 2012
|
|
3TKM
 
 | | Crystal structure PPAR delta binding GW0742 | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor delta, {4-[({2-[3-fluoro-4-(trifluoromethyl)phenyl]-4-methyl-1,3-thiazol-5-yl}methyl)sulfanyl]-2-methylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Batista, F.H, Polikarpov, I. | | Deposit date: | 2011-08-27 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural Insights into Human Peroxisome Proliferator Activated Receptor Delta (PPAR-Delta) Selective Ligand Binding.
Plos One, 7, 2012
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KJO
 
 | |
5KJQ
 
 | | X-ray structure of PcCel45A in complex with cellobiose expressed in Aspergillus nidullans | | Descriptor: | Endoglucanase V-like protein, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Godoy, A.S, Ramia, M.P, Camilo, C.M, Polikarpov, I. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structure, computational and biochemical analysis of PcCel45A endoglucanase from Phanerochaete chrysosporium and catalytic mechanisms of GH45 subfamily C members.
Sci Rep, 8, 2018
|
|
5KSQ
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KSS
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-Ds (Dimer Smaller) | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, A.M.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KST
 
 | | Stationary phase Survival protein E (SurE) from Xylella fastidiosa- XfSurE-TSAmp (Tetramer Smaller - crystallization with 3'AMP). | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
1SZN
 
 | | THE STRUCTURE OF ALPHA-GALACTOSIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brando Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-06 | | Release date: | 2004-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism.
J.Mol.Biol., 339, 2004
|
|
1T0O
 
 | | The structure of alpha-galactosidase from Trichoderma reesei complexed with beta-D-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brandao Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-12 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism
J.Mol.Biol., 339, 2004
|
|
3EMY
 
 | | Crystal structure of Trichoderma reesei aspartic proteinase complexed with pepstatin A | | Descriptor: | Pepstatin, Trichoderma reesei Aspartic protease | | Authors: | Nascimento, A.S, Krauchenco, S, Golubev, A.M, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Statistical coupling analysis of aspartic proteinases based on crystal
structures of the Trichoderma reesei enzyme and its complex with pepstatin A.
J.Mol.Biol., 382, 2008
|
|
3CFT
 
 | |
3CFM
 
 | |
3DLQ
 
 | | Crystal structure of the IL-22/IL-22R1 complex | | Descriptor: | Interleukin-22, Interleukin-22 receptor subunit alpha-1 | | Authors: | Bleicher, L, de Moura, P.R, Watanabe, L, Colau, D, Dumoutier, L, Renauld, J.-C, Polikarpov, I. | | Deposit date: | 2008-06-28 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism
Febs Lett., 582, 2008
|
|
