7T6W
 
 | | Crystal structure of Chaetomium Glucosidase I (apo) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, GLYCEROL, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
7T8V
 
 | | Co-crystal structure of Chaetomium glucosidase I with EB-0159 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
7T66
 
 | | Co-crystal structure of Chaetomium glucosidase with compound UV-4 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
7T68
 
 | | Co-crystal structure of Chaetomium glucosidase with compound UV-5 | | Descriptor: | (2R,3R,4R,5S)-1-[6-(4-azido-2-nitroanilino)hexyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
6VRN
 
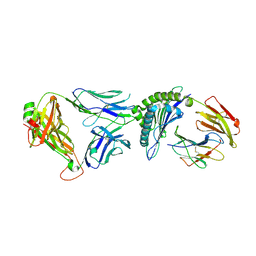 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
6VTC
 
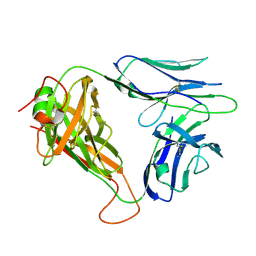 | | p53-specific T cell receptor | | Descriptor: | T-cell Receptor 1a2, p53-specific T cell receptor, B-chain | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
3OWE
 
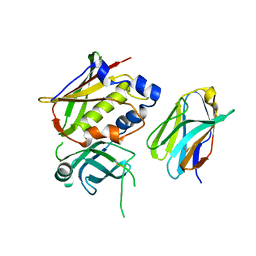 | | Crystal Structure of Staphylococcal Enterotoxin G (SEG) in Complex with a High Affinity Mutant Mouse T-cell Receptor Chain | | Descriptor: | Beta-chain, Enterotoxin SEG | | Authors: | Fernandez, M.M, Cho, S, Robinson, H, Mariuzza, R.A, Malchiodi, M.L. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of staphylococcal enterotoxin G (SEG) in complex with a mouse T-cell receptor {beta} chain.
J.Biol.Chem., 286, 2011
|
|
2IAM
 
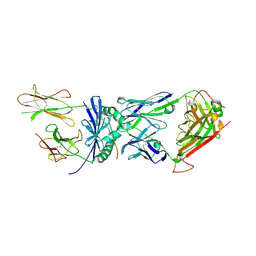 | | Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR | | Descriptor: | 15-mer peptide from Triosephosphate isomerase, CD4+ T cell receptor E8 alpha chain, CD4+ T cell receptor E8 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Mariuzza, R.A. | | Deposit date: | 2006-09-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Nat.Immunol., 8, 2007
|
|
2IAN
 
 | | Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR | | Descriptor: | 15-mer peptide from Triosephosphate isomerase, CD4+ T cell receptor E8 alpha chain, CD4+ T cell receptor E8 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Mariuzza, R.A. | | Deposit date: | 2006-09-08 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Nat.Immunol., 8, 2007
|
|
3O6F
 
 | | Crystal structure of a human autoimmune TCR MS2-3C8 bound to MHC class II self-ligand MBP/HLA-DR4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-4 beta chain, ... | | Authors: | Yin, Y, Li, Y, Martin, R, Mariuzza, R.A. | | Deposit date: | 2010-07-29 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a TCR with high affinity for self-antigen reveals basis for escape from negative selection.
Embo J., 30, 2011
|
|
2G9H
 
 | | Crystal Structure of Staphylococcal Enterotoxin I (SEI) in Complex with a Human MHC class II Molecule | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HLA class II histocompatibility antigen, ... | | Authors: | Fernandez, M.M, Guan, R, Malchiodi, E.L, Mariuzza, R.A. | | Deposit date: | 2006-03-06 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcal enterotoxin I (SEI) in complex with a human major histocompatibility complex class II molecule.
J.Biol.Chem., 281, 2006
|
|
2IAL
 
 | | Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR | | Descriptor: | CD4+ T cell receptor E8 alpha chain, CD4+ T cell receptor E8 beta chain | | Authors: | Deng, L, Langley, R.J, Mariuzza, R.A. | | Deposit date: | 2006-09-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Nat.Immunol., 8, 2007
|
|
1SBB
 
 | | T-CELL RECEPTOR BETA CHAIN COMPLEXED WITH SUPERANTIGEN SEB | | Descriptor: | PROTEIN (14.3.D T CELL ANTIGEN RECEPTOR), PROTEIN (STAPHYLOCOCCAL ENTEROTOXIN B) | | Authors: | Li, H, Mariuzza, R.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the complex between a T cell receptor beta chain and the superantigen staphylococcal enterotoxin B.
Immunity, 9, 1998
|
|
1SK4
 
 | | crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | Peptidoglycan recognition protein I-alpha, SODIUM ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
1R1S
 
 | |
1R1P
 
 | |
1TWQ
 
 | | Crystal structure of the C-terminal PGN-binding domain of human PGRP-Ialpha in complex with PGN analog muramyl tripeptide | | Descriptor: | N-acetyl-beta-muramic acid, NICKEL (II) ION, muramyl tripeptide, ... | | Authors: | Guan, R, Roychowdury, A, Boons, G.-A, Mariuzza, R.A. | | Deposit date: | 2004-07-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for peptidoglycan binding by peptidoglycan recognition proteins
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SK3
 
 | | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | NICKEL (II) ION, Peptidoglycan recognition protein I-alpha, SULFATE ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
7R6J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 1 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[3-methyl-5-(pyrimidin-2-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7REV
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 3 | | Descriptor: | (2R,3R,4R,5S)-1-[(4-{[4-(furan-2-yl)-2-methylanilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
3C8J
 
 | |
7K9N
 
 | | Co-crystal structure of alpha glucosidase with compound 2 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(9-methoxynonyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9O
 
 | | Co-crystal structure of alpha glucosidase with compound 3 | | Descriptor: | (1~{S},2~{S},3~{R},4~{S},5~{S})-1-(hydroxymethyl)-5-[6-[[2-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]-4-(1,2,3,4-tetrazol-1-yl)phenyl]amino]hexylamino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9T
 
 | | Co-crystal structure of alpha glucosidase with compound 5 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-{[(5Z)-6-{[2-nitro-4-(2H-1,2,3-triazol-2-yl)phenyl]amino}hex-5-en-1-yl]amino}cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9Q
 
 | | Co-crystal structure of alpha glucosidase with compound 4 | | Descriptor: | (1S,2S,3R,4S,5S)-5-amino-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
