7MQT
 
 | |
7O2Y
 
 | |
7O3K
 
 | |
7P2B
 
 | |
7Q02
 
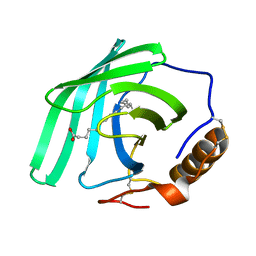 | | Zn-free structure of lipocalin-like Milk protein, inspired from Diploptera punctata, expressed in Saccharomyces cerevisiae | | Descriptor: | Milk protein, PALMITOLEIC ACID | | Authors: | Banerjee, S, Dhanabalan, K.V, Ramaswamy, S. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of recombinantly expressed cockroach Lili-Mip protein in glycosylated and deglycosylated forms.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
6JKU
 
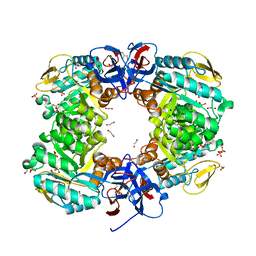 | | Crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella Multocida | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ... | | Authors: | Manjunath, L, Bose, S, Subramanian, R. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Quaternary variations in the structural assembly of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella multocida.
Proteins, 2020
|
|
6XT2
 
 | | EQADH-NADH-HEPTAFLUOROBUTANOL, P21 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,2,3,3,4,4,4-heptafluorobutan-1-ol, ... | | Authors: | Plapp, B.V, Ramaswamy, S. | | Deposit date: | 2020-07-16 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Alternative binding modes in abortive NADH-alcohol complexes of horse liver alcohol dehydrogenase.
Arch.Biochem.Biophys., 701, 2021
|
|
4X1G
 
 | | Crystal structure of the hPXR-LBD in complex with the synthetic estrogen 17alpha-ethinylestradiol and the pesticide trans-nonachlor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Ethinyl estradiol, ISOPROPYL ALCOHOL, ... | | Authors: | Delfosse, V, Huet, T, Bourguet, W. | | Deposit date: | 2014-11-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds.
Nat Commun, 6, 2015
|
|
4X1F
 
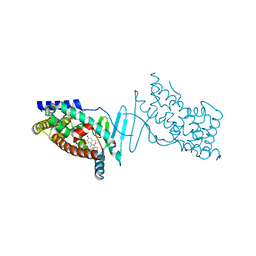 | | Crystal structure of the hPXR-LBD in complex with the synthetic estrogen 17alpha-ethinylestradiol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Ethinyl estradiol, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Delfosse, V, Huet, T, Bourguet, W. | | Deposit date: | 2014-11-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds.
Nat Commun, 6, 2015
|
|
6ON7
 
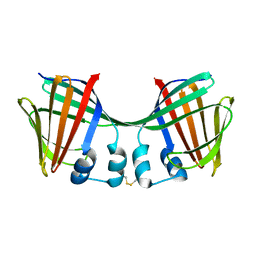 | |
6ON5
 
 | |
6ON8
 
 | |
4XAO
 
 | | Crystal structure of the hPXR-LBD obtained in presence of the pesticide trans-nonachlor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Delfosse, V, Huet, T, Bourguet, W. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds.
Nat Commun, 6, 2015
|
|
6OL9
 
 | | Structure of the M5 muscarinic acetylcholine receptor (M5-T4L) bound to tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Vuckovic, Z, Christopoulos, A, Thal, D.M. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Crystal structure of the M5muscarinic acetylcholine receptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5KJ1
 
 | | G173A horse liver alcohol dehydrogenase complexed with NAD+ and pentafluorobenzyl alcohol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Dependence of crystallographic atomic displacement parameters on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6E50
 
 | |
6E5E
 
 | |
6E5Q
 
 | |
6E7M
 
 | |
6E5S
 
 | |
6B8F
 
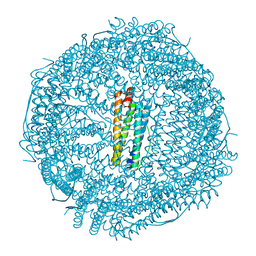 | | Contracted Human Heavy-Chain Ferritin Crystal-Hydrogel Hybrid | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain | | Authors: | Zhang, L, Bailey, J.B, Subramanian, R, Tezcan, F.A. | | Deposit date: | 2017-10-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Hyperexpandable, self-healing macromolecular crystals with integrated polymer networks.
Nature, 557, 2018
|
|
6E6L
 
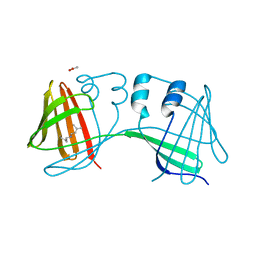 | |
6B8G
 
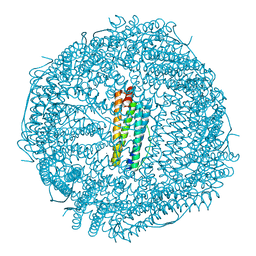 | | Twice-Contracted Human Heavy-Chain Ferritin Crystal-Hydrogel Hybrid | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain | | Authors: | Zhang, L, Bailey, J.B, Subramanian, R, Tezcan, F.A. | | Deposit date: | 2017-10-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Hyperexpandable, self-healing macromolecular crystals with integrated polymer networks.
Nature, 557, 2018
|
|
6E51
 
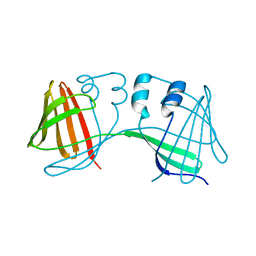 | |
6E5R
 
 | |
