4TWM
 
 | | Crystal structure of dioscorin from Dioscorea japonica | | Descriptor: | Dioscorin 5, SULFATE ION | | Authors: | Xue, Y.L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate Reductase
Mol Plant, 8, 2015
|
|
5YP4
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, LYSINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP1
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP2
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | Descriptor: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP3
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, ISOLEUCINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
4WFS
 
 | | Crystal Structure of tRNA-dihydrouridine(20) synthase catalytic domain | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
5XCY
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium at 1.2 angstrom | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6L96
 
 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | Authors: | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
3FKU
 
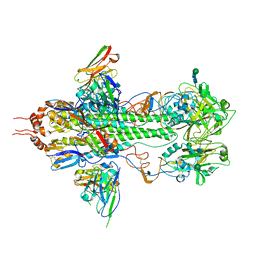 | | Crystal structure of influenza hemagglutinin (H5) in complex with a broadly neutralizing antibody F10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Hwang, W.C, Santelli, E, Stec, B, Wei, G, Cadwell, G, Bankston, L.A, Sui, J, Perez, S, Aird, D, Chen, L.M, Ali, M, Murakami, A, Yammanuru, A, Han, T, Cox, N, Donis, R.O, Liddington, R.C, Marasco, W.A. | | Deposit date: | 2008-12-17 | | Release date: | 2009-02-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5DNW
 
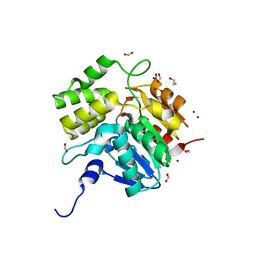 | | Crystal structure of KAI2-like protein from Striga (apo state 1) | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
5DNU
 
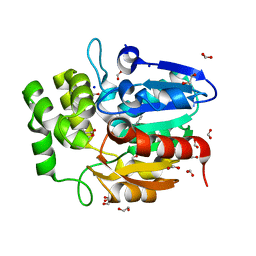 | | Crystal structure of Striga KAI2-like protein in complex with karrikin | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2H-furo[2,3-c]pyran-2-one, BENZOIC ACID, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
5DNV
 
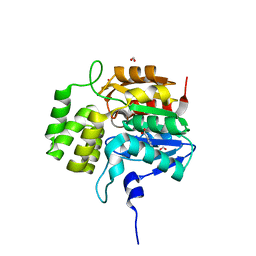 | | Crystal structure of KAI2-like protein from Striga (apo state 2) | | Descriptor: | BENZOIC ACID, FORMIC ACID, ShKAI2iB | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica
Sci Rep, 6, 2016
|
|
5GMT
 
 | |
6JTB
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with citrate from Porphyromonas gingivalis (Space) | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
6JTC
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with SH-5 from Porphyromonas gingivalis (Space) | | Descriptor: | 2-(2-azanylethylamino)-5-nitro-benzoic acid, Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
3K8U
 
 | | Crystal Structure of the Peptidase Domain of Streptococcus ComA, a Bi-functional ABC Transporter Involved in Quorum Sensing Pathway | | Descriptor: | Putative ABC transporter, ATP-binding protein ComA | | Authors: | Ishii, S, Yano, T, Ebihara, A, Okamoto, A, Manzoku, M, Hayashi, H. | | Deposit date: | 2009-10-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP-binding cassette transporter involved in the quorum-sensing pathway
J.Biol.Chem., 285, 2010
|
|
8W96
 
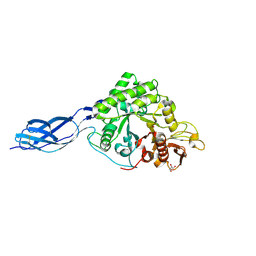 | | SmChiA with diacetyl chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A, GLYCEROL | | Authors: | Tanaka, Y, Nakamura, A. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Product inhibition slow down the moving velocity of processive chitinase and sliding-intermediate state blocks re-binding of product.
Arch.Biochem.Biophys., 752, 2024
|
|
8W8V
 
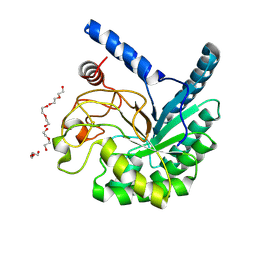 | | High-resolution X-ray structure of cellulase Cel6A from Phanerochaete chrysosporium at cryogenic temperature, Enzyme-Product complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Glucanase, ... | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-09-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4Y
 
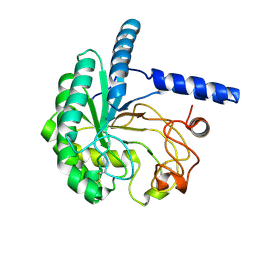 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, low-D2O-solvent | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.4 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4W
 
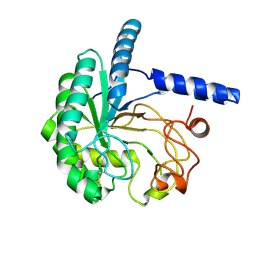 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.36 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W8U
 
 | | High-resolution X-ray structure of cellulase Cel6A from Phanerochaete chrysosporium at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-09-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4X
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, Enzyme-Product complex | | Descriptor: | Glucanase, SODIUM ION, beta-D-glucopyranose, ... | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.4 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4Z
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, Enzyme-Product complex, H2O solvent | | Descriptor: | Glucanase, SODIUM ION, beta-D-glucopyranose, ... | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-30 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
7EDC
 
 | | Crystal structure of mutant tRNA [Gm18] methyltransferase TrmH (E107G) in complex with S-adenosyl-L-methionine from Escherichia coli | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (guanosine(18)-2'-O)-methyltransferase | | Authors: | Kono, Y, Ito, A, Okamoto, A, Yamagami, R, Hirata, A, Hori, H. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Unique substrate specificity of type II tRNA Gm18 methyltransferase from Escherichia coli
To Be Published
|
|
5Y33
 
 | |
