3E83
 
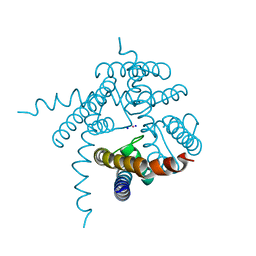 | | Crystal Structure of the the open NaK channel pore | | Descriptor: | CESIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Jiang, Y, Alam, A. | | Deposit date: | 2008-08-19 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of ion selectivity in the NaK channel
Nat.Struct.Mol.Biol., 16, 2009
|
|
7MHY
 
 | |
7MHZ
 
 | |
8JC4
 
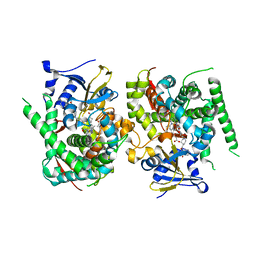 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-Pid-Phe and hydroxylamine | | Descriptor: | 1-pyridin-4-ylpiperidine-4-carboxylic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
8JC3
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Pyd-N-C4-Phe and hydroxylamine | | Descriptor: | 4-(pyridin-4-ylamino)butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2023-05-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C(sp3)-H hydroxylation of Broad-Spectrum Alkanes Catalyzed by an Artificial P450 Peroxygenase Driven by omega-Pyridyl Fatty Acyl Amino Acids.
Mol Catal, 550, 2023
|
|
8HOR
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4CH3)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4CH3)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOS
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4NO2)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4NO2)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOQ
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4CF3)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4CF3)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOU
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Im-N-C4-Phe-Phe | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-N-C4-Phe-Phe, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOP
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Nap-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Nap-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HON
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Tyr-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Tyr-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7EGN
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | H-Bonding Networks Dictate the Molecular Mechanism of H2O2 Activation by P450
Acs Catalysis, 11, 2021
|
|
7CM5
 
 | | Full-length Sarm1 in a self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
5GG4
 
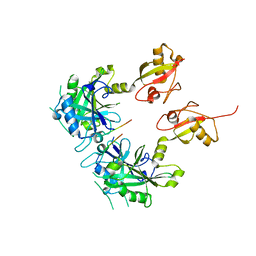 | | Crystal structure of USP7 with RNF169 peptide | | Descriptor: | Peptide from E3 ubiquitin-protein ligase RNF169, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Jiang, Y, Gong, Q. | | Deposit date: | 2016-06-15 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Dual-utility NLS drives RNF169-dependent DNA damage responses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7CM7
 
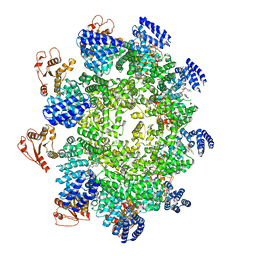 | | NAD+-bound Sarm1 E642A in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM6
 
 | | NAD+-bound Sarm1 in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
3V5U
 
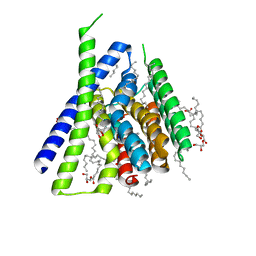 | | Structure of Sodium/Calcium Exchanger from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ACETATE ION, CALCIUM ION, ... | | Authors: | Jiang, Y, Liao, J, Li, H, Zeng, W, Sauer, D, Belmares, R. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Science, 335, 2012
|
|
8HGC
 
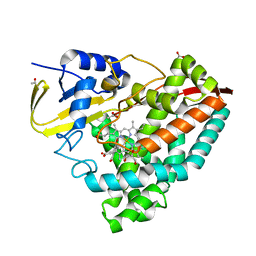 | |
8HGT
 
 | |
6A1K
 
 | | Phosphate acyltransferase PlsX from B.subtilis | | Descriptor: | Phosphate acyltransferase, SULFATE ION | | Authors: | Guo, Z, Jiang, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of an amphipathic peptide sensor of the Bacillus subtilisfluid membrane microdomains.
Commun Biol, 2, 2019
|
|
7WDH
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WDG
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7Y0T
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanyl-L-phenylalanine | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, I7X-PHE-PHE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-06-06 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3LYO
 
 | | CROSS-LINKED CHICKEN LYSOZYME CRYSTAL IN 95% ACETONITRILE-WATER | | Descriptor: | ACETONITRILE, LYSOZYME | | Authors: | Huang, Q, Wang, Z, Zhu, G, Qian, M, Shao, M, Jia, Y, Tang, Y. | | Deposit date: | 1998-03-11 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray studies on cross-linked lysozyme crystals in acetonitrile-water mixture.
Biochim.Biophys.Acta, 1384, 1998
|
|
1F2S
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN BOVINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY AT 1.8 A RESOLUTION | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR A | | Authors: | Zhu, Y, Huang, Q, Qian, M, Jia, Y, Tang, Y. | | Deposit date: | 2000-05-29 | | Release date: | 2000-06-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex formed between bovine beta-trypsin and MCTI-A, a trypsin inhibitor of squash family, at 1.8-A resolution.
J.Protein Chem., 18, 1999
|
|
