7W9B
 
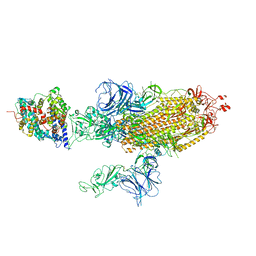 | | SARS-CoV-2 Delta S-ACE2-C2b | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9F
 
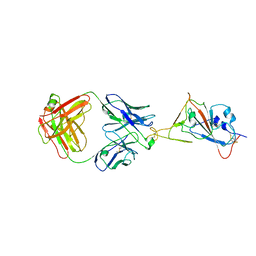 | | SARS-CoV-2 Delta S-RBD-8D3 | | Descriptor: | Spike protein S1, The heavy chain of 8D3, The light chain of 8D3 | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9I
 
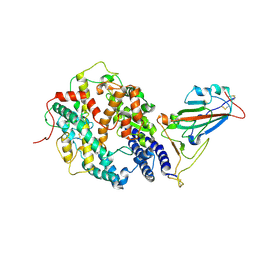 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7WK3
 
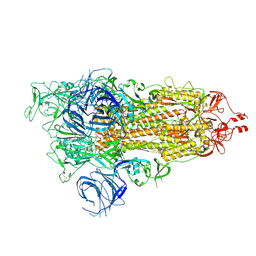 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
4WY4
 
 | | Crystal structure of autophagic SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Zhao, M, Brunger, A.T. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes.
Nature, 520, 2015
|
|
7Y4F
 
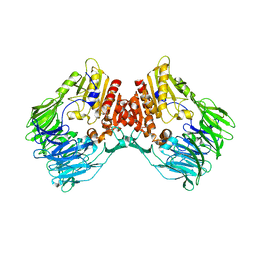 | | bacterial DPP4 | | Descriptor: | Dipeptidyl peptidase IV | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
7Y4G
 
 | | sit-bound btDPP4 | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
4YVU
 
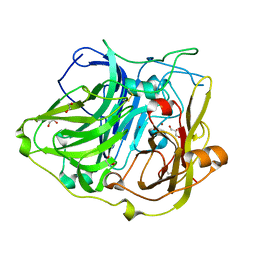 | | Crystal structure of CotA native enzyme in the acid condition, PH5.6 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2015-03-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel binding site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4YVN
 
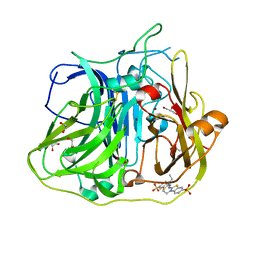 | | Crystal structure of CotA laccase complexed with ABTS at a novel binding site | | Descriptor: | 1,2-ETHANEDIOL, 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, COPPER (II) ION, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2015-03-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel binding site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6LBS
 
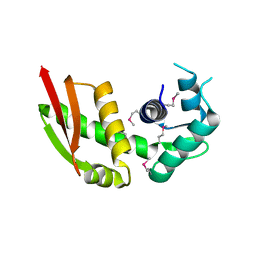 | | Crystal structure of yeast Stn1 | | Descriptor: | KLLA0C11825p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBR
 
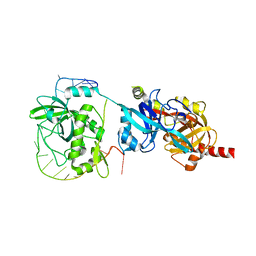 | | Crystal structure of yeast Cdc13 and ssDNA | | Descriptor: | KLLA0F20922p, Telomere single-strand DNA | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBT
 
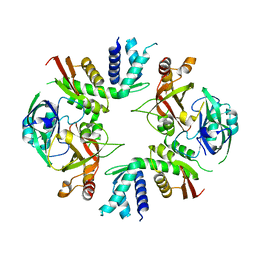 | | Crystal structure of yeast Cdc13 and Stn1 | | Descriptor: | GLYCEROL, KLLA0C11825p, KLLA0F20922p,KLLA0F20922p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBU
 
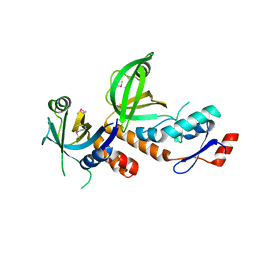 | | Crystal structure of yeast Stn1 and Ten1 | | Descriptor: | KLLA0C11825p, KLLA0E09417p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7L5S
 
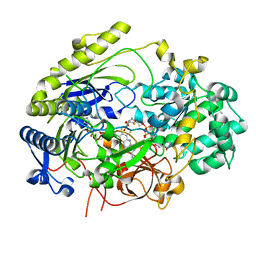 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 5.5 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7L5I
 
 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 7.0 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
7BR8
 
 | | Epstein-Barr virus, C5 penton vertex, CATC absent. | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BSI
 
 | |
7BQT
 
 | | Epstein-Barr virus, C12 portal dodecamer | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BQX
 
 | | Epstein-Barr virus, C5 portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BR7
 
 | | Epstein-Barr virus, C1 portal-proximal penton vertex, CATC binding | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
6JYJ
 
 | | Crystal structure of FAM46B (TENT5B) | | Descriptor: | CITRATE ANION, Terminal nucleotidyltransferase 5B | | Authors: | Zhang, H, Hu, J.L, Gao, S. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.693199 Å) | | Cite: | FAM46B is a prokaryotic-like cytoplasmic poly(A) polymerase essential in human embryonic stem cells.
Nucleic Acids Res., 48, 2020
|
|
7EMN
 
 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
8HAY
 
 | | d4-bound btDPP4 | | Descriptor: | (1~{R})-1-[[4-[5-[[(1~{R})-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]methyl]-2-methoxy-phenoxy]phenyl]methyl]-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinoline, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-10-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
5YD0
 
 | | Crystal structure of Schlafen 13 (SLFN13) N'-domain | | Descriptor: | Schlafen 8, ZINC ION | | Authors: | Yang, J.-Y, Gao, S. | | Deposit date: | 2017-09-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.182 Å) | | Cite: | Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control
Nat Commun, 9, 2018
|
|
