5H83
 
 | | HETEROYOHIMBINE SYNTHASE HYS FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase HYS | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
7NWP
 
 | | A carbohydrate binding module family 9 (CBM9) from Caldicellulosiruptor kristjansonii in complex with cellobiose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CALCIUM ION, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
4YXX
 
 | |
3OJN
 
 | | Structure of Mn-substituted Homoprotocatechuate 2,3-Dioxygenase at 1.65 Ang resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Fielding, A.J, Kovaleva, E.G, Farquhar, E.R, Lipscomb, J.D, Que Jr, L. | | Deposit date: | 2010-08-23 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase.
J.Biol.Inorg.Chem., 16, 2011
|
|
5FX6
 
 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | RHINOVIRUS 3C PROTEASE, ethyl (4R)-4-[[(2S,4S)-1-[(2S)-3-methyl-2-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]butanoyl]-4-phenyl-pyrrolidin-2-yl]carbonylamino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5FSA
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from a pathogenic yeast Candida albicans in complex with the antifungal drug posaconazole | | Descriptor: | CYP51 VARIANT1, POSACONAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Friggeri, L, Lepesheva, G.I. | | Deposit date: | 2016-01-02 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural analyses of Candida albicans sterol 14 alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis.
J. Biol. Chem., 292, 2017
|
|
5AK6
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 2-[(5-CHLORO-2-PYRIDYL)SULFANYL]ETHANOL, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
7NEC
 
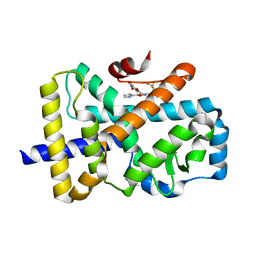 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM217 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-2-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
5FKE
 
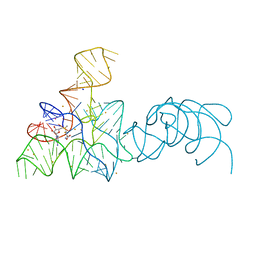 | |
5G0M
 
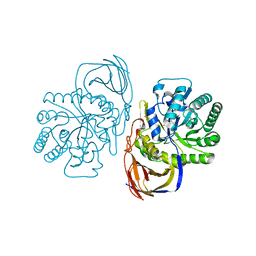 | | beta-glucuronidase with an activity-based probe (N-acyl cyclophellitol aziridine) bound | | Descriptor: | (1S,2R,3R,4S,6S)-6-[(8-azidooctanoyl)amino]-2,3,4-trihydroxycyclohexane-1-carboxylate, BETA-GLUCURONIDASE, PHOSPHATE ION | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a Bacteria Beta-Glucuronidase in Gh79 Family
To be Published
|
|
5FXY
 
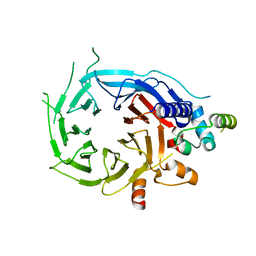 | | Structure of the human RBBP4:MTA1(464-546) complex | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, METASTASIS-ASSOCIATED PROTEIN MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
2J8A
 
 | | X-ray structure of the N-terminus RRM domain of Set1 | | Descriptor: | HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC | | Authors: | Tresaugues, L, Dehe, P.M, Guerois, R, Rodriguez-Gil, A, Varlet, I, Salah, P, Pamblanco, M, Luciano, P, Quevillon-Cheruel, S, Sollier, J, Leulliot, N, Couprie, J, Tordera, V, Zinn-Justin, S, Chavez, S, Van Tilbeurgh, H, Geli, V. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structure of the N-Terminus Rrm Domain of Set1
To be Published
|
|
5FK5
 
 | |
5HAQ
 
 | | OXa-48 beta-lactamase mutant - S70G | | Descriptor: | Beta-lactamase, CADMIUM ION, FORMIC ACID | | Authors: | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
7NJE
 
 | | gamma(S)-crystallin 9-site deamidation mutant grown inside HARE serial crystallography chip | | Descriptor: | Gamma-crystallin S | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJH
 
 | | HEX1 (in cellulo) grown inside HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7ND1
 
 | | First-in-class small molecule inhibitors of Polycomb Repressive Complex 1 (PRC1) RING domain | | Descriptor: | 3-(2-chlorophenyl)-4-ethyl-5-(1~{H}-indol-4-yl)-1~{H}-pyrrole-2-carboxylic acid, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1, ... | | Authors: | Cierpicki, T, Lund, G, Jaremko, L. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
7NJF
 
 | | Hen egg white lysozyme (HEWL) grown inside HARE serial crystallography chip | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ACM
 
 | | Mcg immunoglobulin variable domain with methylene blue | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, GLYCEROL, MCG, ... | | Authors: | Brumshtein, B, Esswein, S.R, Salwinski, L, Phillips, M.L, Ly, A.T, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain.
Elife, 4, 2015
|
|
7NUD
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML734 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[3-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]propylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
3DVW
 
 | |
7NUE
 
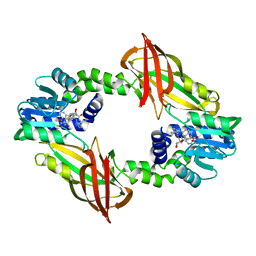 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML736 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
3OJK
 
 | | Structure of Co-substituted Homoprotocatechuate 2,3-Dioxygenase in complex with 4-nitrocatechol at 1.68 Ang resolution | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fielding, A.J, Kovaleva, E.G, Farquhar, E.R, Lipscomb, J.D, Que Jr, L. | | Deposit date: | 2010-08-23 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase.
J.Biol.Inorg.Chem., 16, 2011
|
|
5AI6
 
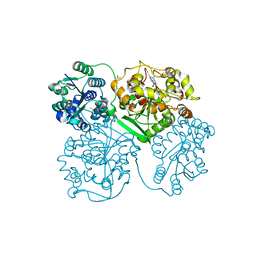 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 5-BROMOQUINOLINE, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
7O14
 
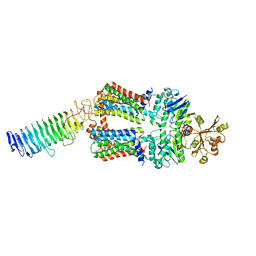 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 1 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
