8IHN
 
 | | Cryo-EM structure of the Rpd3S core complex | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
6IS3
 
 | |
7D14
 
 | | Mouse KCC2 | | Descriptor: | Solute carrier family 12 member 5 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7D10
 
 | | Human NKCC1 | | Descriptor: | PALMITIC ACID, Solute carrier family 12 member 2 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-12 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
1Y4K
 
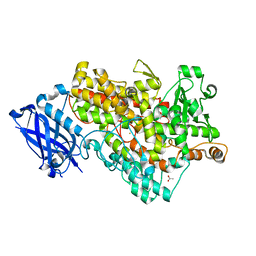 | | Lipoxygenase-1 (Soybean) at 100K, N694G Mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (II) ION, ... | | Authors: | Chruszcz, M, Segraves, E, Holman, T.R, Minor, W. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic, spectroscopic, and structural investigations of the soybean lipoxygenase-1 first-coordination sphere mutant, Asn694Gly.
Biochemistry, 45, 2006
|
|
2KJ1
 
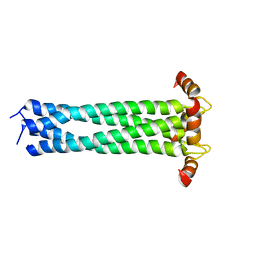 | |
2KIX
 
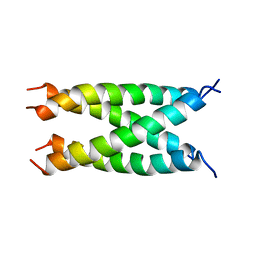 | |
1ZA6
 
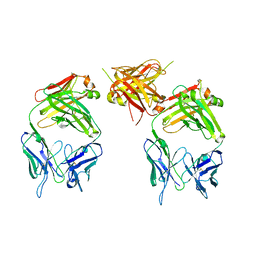 | | The structure of an antitumor CH2-domain-deleted humanized antibody | | Descriptor: | IGG Heavy chain, IGG Light chain | | Authors: | Larson, S.B, Day, J.S, Glaser, S, Braslawsky, G, McPherson, A. | | Deposit date: | 2005-04-05 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of an Antitumor C(H)2-domain-deleted Humanized Antibody.
J.Mol.Biol., 348, 2005
|
|
2OCG
 
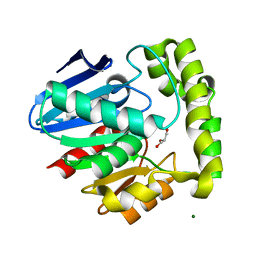 | | Crystal structure of human valacyclovir hydrolase | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2ONO
 
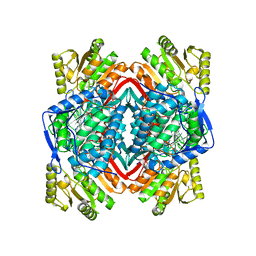 | |
2OCI
 
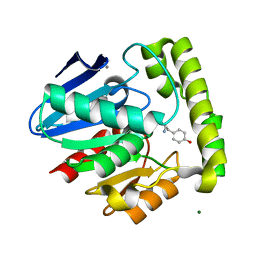 | | Crystal structure of valacyclovir hydrolase complexed with a product analogue | | Descriptor: | L-TYROSINAMIDE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2OCK
 
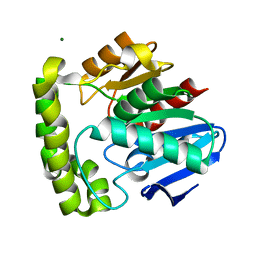 | | Crystal structure of valacyclovir hydrolase D123N mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Valacyclovir hydrolase | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2OCL
 
 | | Crystal structure of valacyclovir hydrolase S122A mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Valacyclovir hydrolase | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2ONM
 
 | | Human Mitochondrial Aldehyde Dehydrogenase Asian Variant, ALDH2*2, complexed with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, ... | | Authors: | Larson, H.N, Hurley, T.D. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487.
J.Biol.Chem., 282, 2007
|
|
2ONN
 
 | |
2ONP
 
 | | Arg475Gln Mutant of Human Mitochondrial Aldehyde Dehydrogenase, complexed with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, GUANIDINE, ... | | Authors: | Larson, H.N, Hurley, T.D. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487.
J.Biol.Chem., 282, 2007
|
|
4N5X
 
 | |
1THA
 
 | | MECHANISM OF MOLECULAR RECOGNITION. STRUCTURAL ASPECTS OF 3,3'-DIIODO-L-THYRONINE BINDING TO HUMAN SERUM TRANSTHYRETIN | | Descriptor: | 3,3'-DEIODO-THYROXINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Luft, J, Cody, V. | | Deposit date: | 1991-11-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of molecular recognition. Structural aspects of 3,3'-diiodo-L-thyronine binding to human serum transthyretin.
J.Biol.Chem., 267, 1992
|
|
2GJ4
 
 | | Structure of rabbit muscle glycogen phosphorylase in complex with ligand | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-CHLORO-N-[(1R,2R)-1-HYDROXY-2,3-DIHYDRO-1H-INDEN-2-YL]-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Otterbein, L.R, Pannifer, A.D, Tucker, J, Breed, J, Oikonomakos, N.G, Rowsell, S, Pauptit, R.A, Claire, M. | | Deposit date: | 2006-03-30 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel thienopyrrole glycogen phosphorylase inhibitors: synthesis, in vitro SAR and crystallographic studies.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1TLM
 
 | |
2GM9
 
 | | Structure of rabbit muscle glycogen phosphorylase in complex with thienopyrrole | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-CHLORO-N-[(3R)-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-3-YL]-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Otterbein, L.R, Pannifer, A.D, Tucker, J, Breed, J, Oikonomakos, N.G, Minshull, C, Rowsell, S, Pauptit, R.A. | | Deposit date: | 2006-04-06 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel thienopyrrole glycogen phosphorylase inhibitors: synthesis, in vitro SAR and crystallographic studies.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1B6V
 
 | | CRYSTAL STRUCTURE OF A HYBRID BETWEEN RIBONUCLEASE A AND BOVINE SEMINAL RIBONUCLEASE | | Descriptor: | RIBONUCLEASE | | Authors: | Vatzaki, E.H, Allen, S.C, Leonidas, D.D, Acharya, K.R. | | Deposit date: | 1999-01-18 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a hybrid between ribonuclease A and bovine seminal ribonuclease--the basic surface, at 2.0 A resolution.
Eur.J.Biochem., 260, 1999
|
|
2PAB
 
 | |
1X32
 
 | |
1THC
 
 | | CRYSTAL STRUCTURE DETERMINATION AT 2.3A OF HUMAN TRANSTHYRETIN-3',5'-DIBROMO-2',4,4',6-TETRA-HYDROXYAURONE COMPLEX | | Descriptor: | 3',5'-DIBROMO-2',4,4',6'-TETRAHYDROXY AURONE, TRANSTHYRETIN | | Authors: | Ciszak, E, Cody, V, Luft, J.R. | | Deposit date: | 1992-04-20 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure determination at 2.3-A resolution of human transthyretin-3',5'-dibromo-2',4,4',6-tetrahydroxyaurone complex.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
