8G6E
 
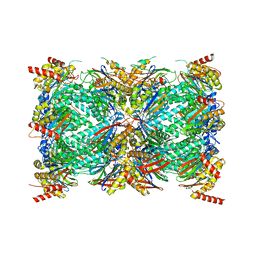 | | Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8G6F
 
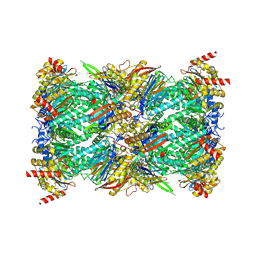 | | Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, Proteasome endopeptidase complex, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
1R5I
 
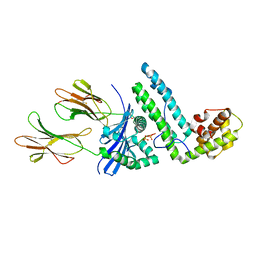 | | Crystal structure of the MAM-MHC complex | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Zhao, Y, Li, Z, Drozd, S.J, Guo, Y, Mourad, W, Li, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Mycoplasma arthritidis mitogen complexed with HLA-DR1 reveals a novel superantigen fold and a dimerized superantigen-MHC complex.
Structure, 12, 2004
|
|
7RD6
 
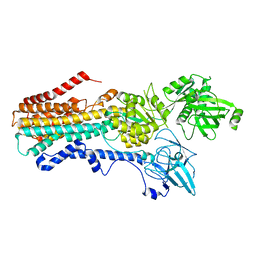 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD8
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E1-ATP state | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.64 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD7
 
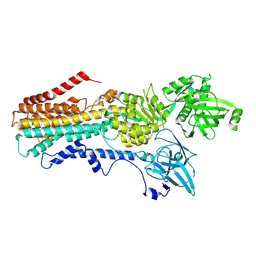 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P-transition state | | Descriptor: | MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1, TETRAFLUOROALUMINATE ION | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RQH
 
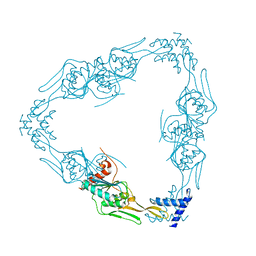 | |
7RPQ
 
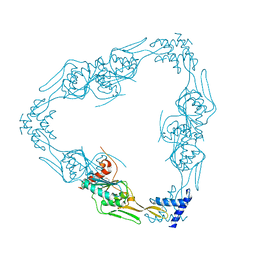 | |
7RQF
 
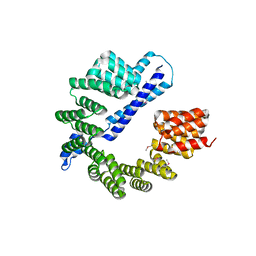 | |
7SUK
 
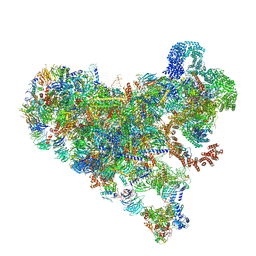 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | Descriptor: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-06 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
8SXG
 
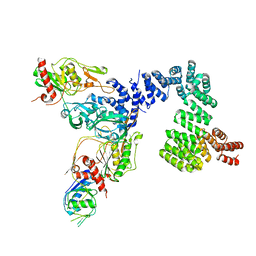 | |
8SXE
 
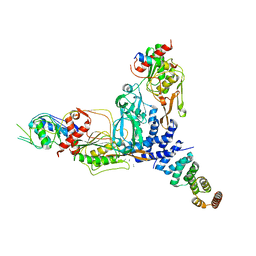 | |
8SXH
 
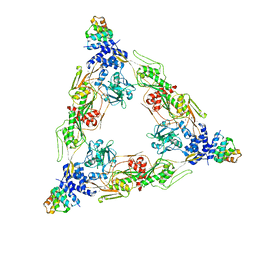 | |
8SXF
 
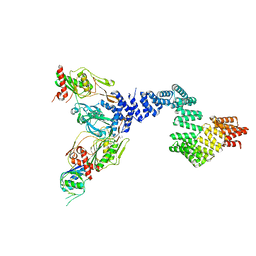 | |
1D1X
 
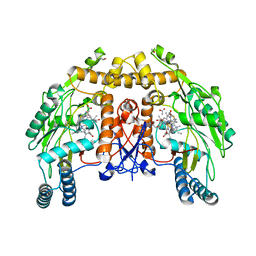 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 1,4-PBITU (H4B BOUND) | | Descriptor: | 2-{2-[4-(2-CARBAMIMIDOYLSULFANYL-ETHYL)-PHENYL]-ETHYL}-ISOTHIOUREA, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
1D1V
 
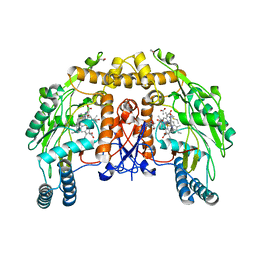 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH S-ETHYL-N-PHENYL-ISOTHIOUREA (H4B BOUND) | | Descriptor: | 2-ETHYL-1-PHENYL-ISOTHIOUREA, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
1D1Y
 
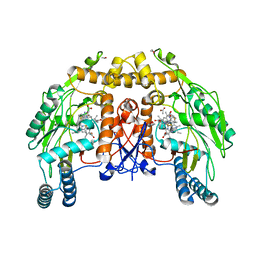 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 1,3-PBITU (H4B FREE) | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[3-(2-CARBAMIMIDOYLSULFANYL-ETHYL)-PHENYL]-ETHYL}-ISOTHIOUREA, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
6E15
 
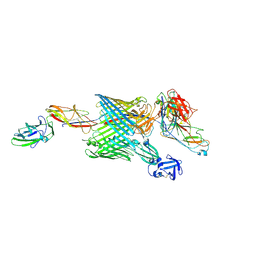 | | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | | Descriptor: | Chaperone protein FimC, Fimbrial biogenesis outer membrane usher protein, Protein FimF, ... | | Authors: | Du, M, Yuan, Z, Yu, H, Henderson, N, Sarowar, S, Zhao, G, Werneburg, G.T, Thanassi, D.G, Li, H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.
Nature, 562, 2018
|
|
6E14
 
 | | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | | Descriptor: | Chaperone protein FimC, Fimbrial biogenesis outer membrane usher protein, Protein FimF, ... | | Authors: | Du, M, Yuan, Z, Yu, H, Henderson, N, Sarowar, S, Zhao, G, Werneburg, G.T, Thanassi, D.G, Li, H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.
Nature, 562, 2018
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
1PKF
 
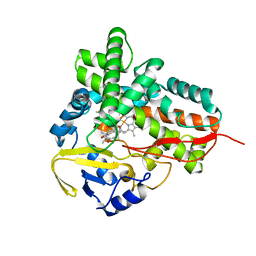 | | Crystal Structure of Epothilone D-bound Cytochrome P450epoK | | Descriptor: | EPOTHILONE D, PROTOPORPHYRIN IX CONTAINING FE, cytochrome p450EpoK | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-06-05 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Epothilone D-bound, Epothilone B-bound, and Substrate-free Forms of Cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
5TGN
 
 | | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus
To Be Published
|
|
1Q5D
 
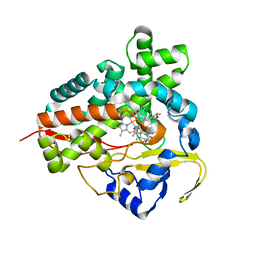 | | Epothilone B-bound Cytochrome P450epoK | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1Q5E
 
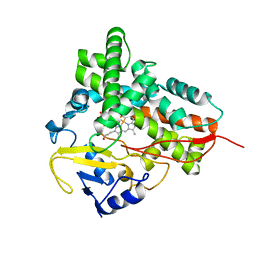 | | Substrate-free Cytochrome P450epoK | | Descriptor: | P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
