7W86
 
 | | Crystal structure of the DYW domain of DYW1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Sawada, Y, Shimizu, H, Toma-Fukai, S, Shimizu, T. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the activation of an Arabidopsis organellar C-to-U RNA editing enzyme by active site complementation.
Plant Cell, 35, 2023
|
|
1IE6
 
 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | Descriptor: | IMPERATOXIN A | | Authors: | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | Deposit date: | 2001-04-07 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
1GIQ
 
 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
1FV3
 
 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH AN ANALOGUE OF ITS GANGLIOSIDE RECEPTOR GT1B | | Descriptor: | ETHYL-TRIMETHYL-SILANE, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Fotinou, C, Emsley, P, Black, I, Ando, H, Ishida, H, Kiso, M, Sinha, K.A, Fairweather, N.F, Isaacs, N.W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin.
J.Biol.Chem., 276, 2001
|
|
8FJN
 
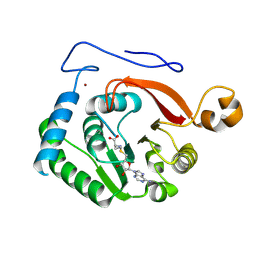 | |
8FJM
 
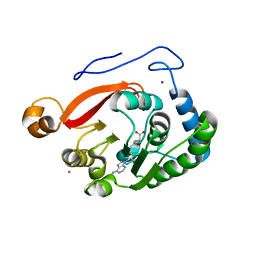 | |
8GSR
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (apo-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8GST
 
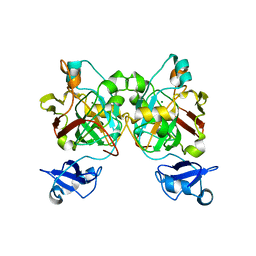 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (pyruvate bound-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION, PYRUVIC ACID | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
6KJG
 
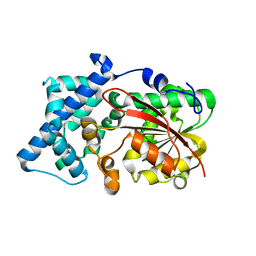 | | Crystal structure of PsoF | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
3Q2Z
 
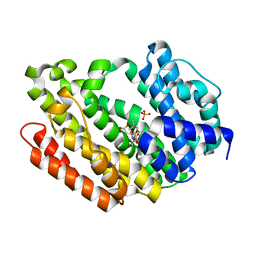 | | Human Squalene synthase in complex with N-[(3R,5S)-7-Chloro-5-(2,3-dimethoxyphenyl)-1-neopentyl-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepine-3-acetyl]-L-aspartic acid | | Descriptor: | N-{[(3R,5S)-7-chloro-5-(2,3-dimethoxyphenyl)-1-(2,2-dimethylpropyl)-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepin-3-yl]acetyl}-L-aspartic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Shimizu, H, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
6KJI
 
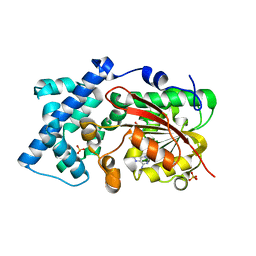 | | Crystal structure of PsoF with SAH | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
7WNK
 
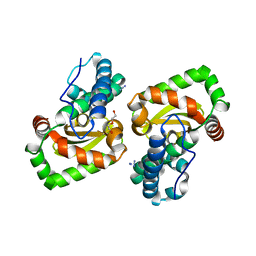 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase K38R and A121E | | Descriptor: | AZIDE ION, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
7WNL
 
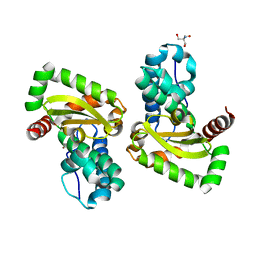 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase K38R and A121Y | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, BROMIDE ION, ... | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
6L96
 
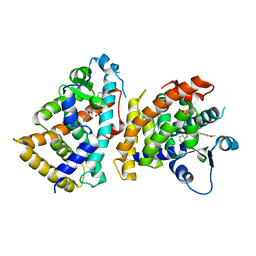 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | Authors: | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
7UPT
 
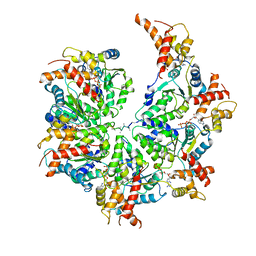 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (open conformation) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | Deposit date: | 2022-04-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
7UPR
 
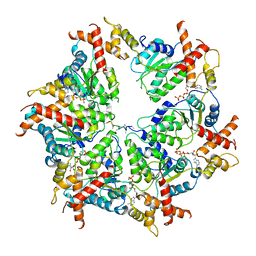 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (closed conformation) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Outer mitochondrial transmembrane helix translocase, ... | | Authors: | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | Deposit date: | 2022-04-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
8HMN
 
 | |
2AHL
 
 | | Crystal structure of the hydroxylamine-induced deoxy-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase in complex with a caddie protein | | Descriptor: | CADDIE PROTEIN ORF378, COPPER (I) ION, NITRATE ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-07-28 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
5YY2
 
 | | Crystal structure of AsqI with Zn | | Descriptor: | Uncharacterized protein AsqI, ZINC ION | | Authors: | Hara, K, Hashimoto, H, Kishimoto, S, Watanabe, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Enzymatic one-step ring contraction for quinolone biosynthesis.
Nat Commun, 9, 2018
|
|
5GHD
 
 | |
5Z1N
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
5Z39
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum form II | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2018-01-05 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
5YY3
 
 | |
