3MZL
 
 | | Sec13/Sec31 edge element, loop deletion mutant | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Whittle, J.R, Schwartz, T.U. | | Deposit date: | 2010-05-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
J.Cell Biol., 190, 2010
|
|
4XVS
 
 | | Crystal structure of HIV-1 donor 45 d45-01dG5 coreE gp120 with antibody 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (VRC07_1995) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 Light chain, Donor 45 01dG5 coreE gp120, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-01-27 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
4CHJ
 
 | |
8C07
 
 | |
8C06
 
 | |
4XK8
 
 | | Crystal structure of plant photosystem I-LHCI super-complex at 2.8 angstrom resolution | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Suga, M, Qin, X, Kuang, T, Shen, J.R. | | Deposit date: | 2015-01-10 | | Release date: | 2015-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex
Science, 348, 2015
|
|
8CH7
 
 | | RDC-refined Interleukin-4 (wild type) pH 5.6 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8CGF
 
 | | Interleukin-4 (wild type) pH 2.4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
4XVT
 
 | | Crystal structure of HIV-1 93TH057 coreE gp120 with antibody 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (VRC07_1995) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (VRC07_1995) Light chain, ENVELOPE GLYCOPROTEIN GP120 OF HIV-1 CLADE A/E, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
3NGG
 
 | | X-ray Structure of Omwaprin | | Descriptor: | Omwaprin-a | | Authors: | Banigan, J.R, Mandal, K, Sawaya, M.R, Thammavongsa, V, Hendrickx, A.P.A, Schneewind, O, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Protein Sci., 19, 2010
|
|
3NTB
 
 | | Structure of 6-methylthio naproxen analog bound to mCOX-2. | | Descriptor: | (2S)-2-[6-(methylsulfanyl)naphthalen-2-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3NT1
 
 | | High resolution structure of naproxen:COX-2 complex. | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3NPL
 
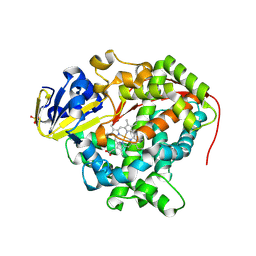 | | Structure of Ru(bpy)2(A-Phen)(K97C) P450 BM3 heme domain, a ruthenium modified P450 BM3 mutant | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Ener, M, Lee, Y.-T, Goodin, D.B, Winkler, J.R, Gray, H.B, Cheruzel, L. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Ru(bpy)2(A-Phen)(K97C) P450 BM3 heme domain, a ruthenium modified P450 BM3 mutant
To be Published
|
|
4DDC
 
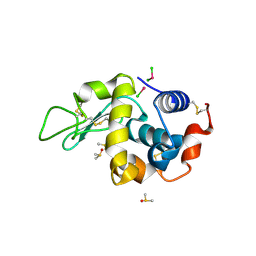 | | EVAL processed HEWL, cisplatin DMSO NAG silicone oil | | Descriptor: | Cisplatin, DIMETHYL SULFOXIDE, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3O2X
 
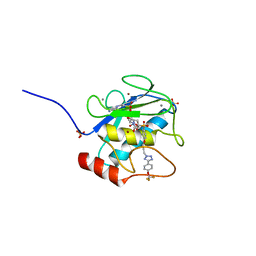 | | MMP-13 in complex with selective tetrazole core inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Collagenase 3, ... | | Authors: | Kiefer, J.R, Barta, T.E, Becker, D.P, Mathis, K.J, Williams, J. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MMP-13 in complex with selective tetrazole core inhibitor
To be Published
|
|
1UA0
 
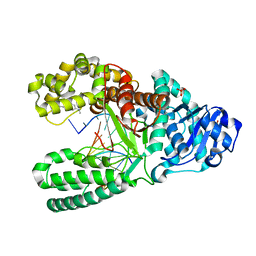 | | Aminofluorene DNA adduct at the pre-insertion site of a DNA polymerase | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
3O8C
 
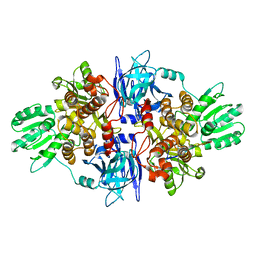 | |
4DD7
 
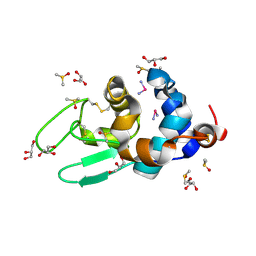 | | EVAL processed HEWL, carboplatin DMSO glycerol | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Lysozyme C, ... | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3O8D
 
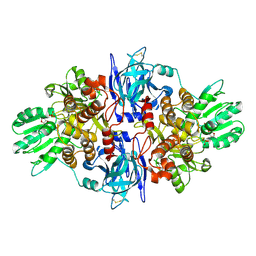 | |
1UA1
 
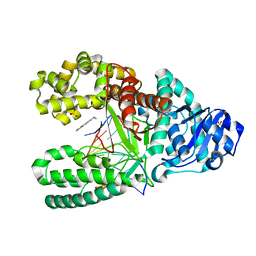 | | Structure of aminofluorene adduct paired opposite cytosine at the polymerase active site. | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
8D1U
 
 | | E. coli beta-ketoacyl-[acyl carrier protein] synthase III (FabH) with an acetylated cysteine and in complex with oxa(dethia)-Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, CHLORIDE ION, oxa(dethia)-CoA | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-ketoacylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(dethia)CoA.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3NRU
 
 | | Ligand binding domain of EPHA7 | | Descriptor: | CHLORIDE ION, Ephrin receptor, SULFATE ION | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2010-06-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ephrin A7 ligand binding domain
To be Published
|
|
8CXL
 
 | | Structure of NapH3, a vanadium-dependent haloperoxidase homolog catalyzing the stereospecific alpha-hydroxyketone rearrangement reaction in napyradiomycin biosynthesis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NapH3 | | Authors: | Chen, P.Y.-T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 61, 2022
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4YBO
 
 | |
