7JOV
 
 | |
1KX8
 
 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
6JBI
 
 | | Structure of Tps1 apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Yi, L, Wang, D, Liu, J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
6JAK
 
 | | OtsA apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-24 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
7MC4
 
 | |
6J0Q
 
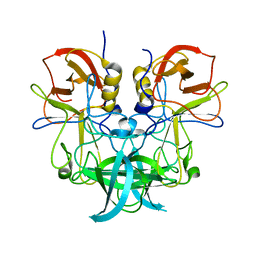 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
4LN2
 
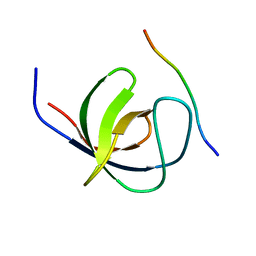 | | The second SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | Descriptor: | Sorbin and SH3 domain-containing protein 1, proline rich peptide | | Authors: | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2013-07-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
5NDF
 
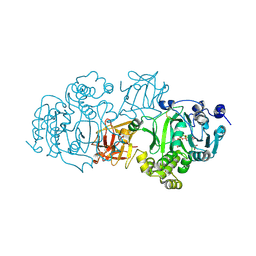 | | Small-molecule inhibition of ppGalNAc-Ts selectively reduces mucin-type O-glycosylation | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, MANGANESE (II) ION, ... | | Authors: | Hurtado-Guerrero, R, De las Rivas, M. | | Deposit date: | 2017-03-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The small molecule luteolin inhibits N-acetyl-alpha-galactosaminyltransferases and reduces mucin-type O-glycosylation of amyloid precursor protein.
J. Biol. Chem., 292, 2017
|
|
7TIK
 
 | |
6JBW
 
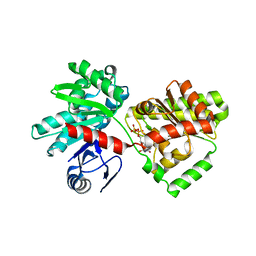 | | Structure of Tps1/UDP complex | | Descriptor: | Trehalose-6-phosphate synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
7R40
 
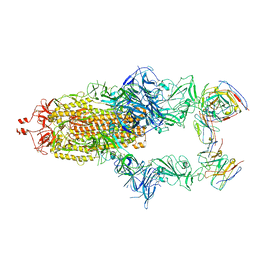 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 87G7 heavy chain variable region, ... | | Authors: | Hurdiss, D.L. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-20 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern.
Sci Immunol, 7, 2022
|
|
1AUV
 
 | |
2OZT
 
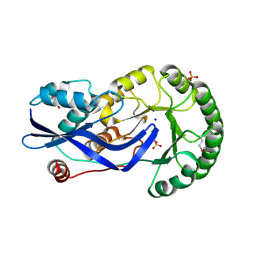 | | Crystal structure of O-succinylbenzoate synthase from Thermosynechococcus elongatus BP-1 | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tlr1174 protein | | Authors: | Malashkevich, V.N, Bonanno, J, Toro, R, Sauder, J.M, Schwinn, K.D, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Gheyi, T, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OLA
 
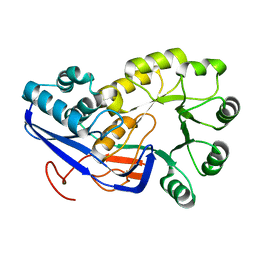 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, cubic crystal form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Sauder, J.M, Ozyurt, S, Wasserman, S.R, Smith, D, Dickey, M, Maletic, M, Reyes, C, Gheyi, T, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CAW
 
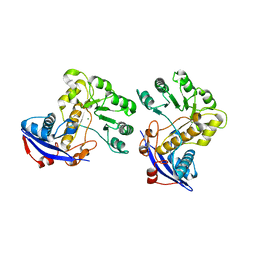 | | Crystal structure of o-succinylbenzoate synthase from Bdellovibrio bacteriovorus liganded with Mg | | Descriptor: | MAGNESIUM ION, o-succinylbenzoate synthase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7U6R
 
 | |
5XSJ
 
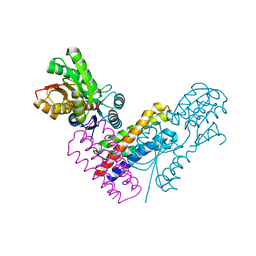 | | XylFII-LytSN complex | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS, ... | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5HXL
 
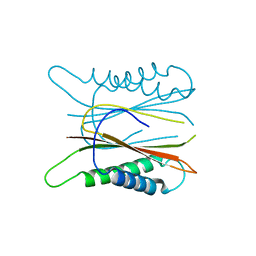 | |
5XSD
 
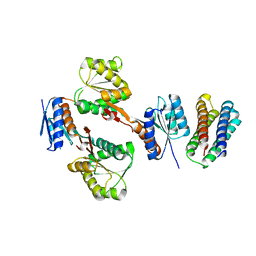 | | XylFII-LytSN complex mutant - D103A | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XSS
 
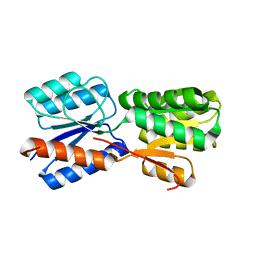 | | XylFII molecule | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, beta-D-xylopyranose | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5HYC
 
 | | Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Uncharacterized protein | | Authors: | Liu, J, Li, G, Huang, J, Peng, Y.-l. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Sci Rep, 8, 2018
|
|
1WUE
 
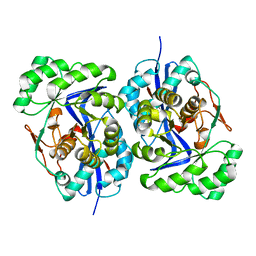 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WUF
 
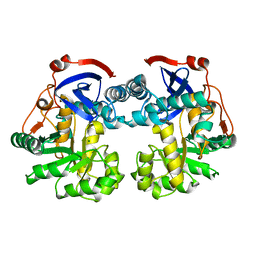 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6UVB
 
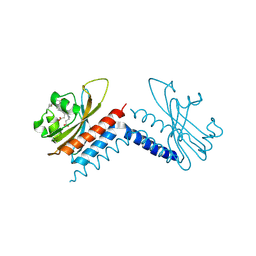 | |
1I38
 
 | |
