7Y5T
 
 | | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5Z
 
 | | CryoEM structure of human PS2-containing gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
3TJ0
 
 | | Crystal Structure of Influenza B Virus Nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Ng, A.K.L, Zhang, H, Liu, J, Au, S.W.N, Wang, J, Shaw, P.C. | | Deposit date: | 2011-08-23 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.233 Å) | | Cite: | Structural basis for RNA binding and homo-oligomer formation by influenza B virus nucleoprotein
J.Virol., 86, 2012
|
|
1NJ1
 
 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to cysteine sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ5
 
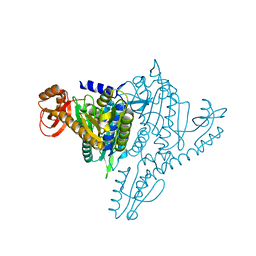 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to proline sulfamoyl adenylate | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ8
 
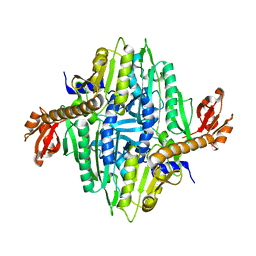 | | Crystal Structure of Prolyl-tRNA Synthetase from Methanocaldococcus janaschii | | Descriptor: | Proline-tRNA Synthetase | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NGM
 
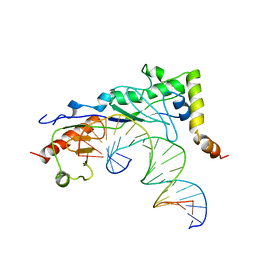 | | Crystal structure of a yeast Brf1-TBP-DNA ternary complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*AP*AP*AP*AP*AP*AP*AP*TP*GP*TP*TP*TP*TP*TP*T)-3', Transcription factor IIIB BRF1 subunit, ... | | Authors: | Juo, Z.S, Kassavetis, G.A, Wang, J, Geiduschek, E.P, Sigler, P.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a transcription factor IIIB core interface ternary complex
Nature, 422, 2003
|
|
1M60
 
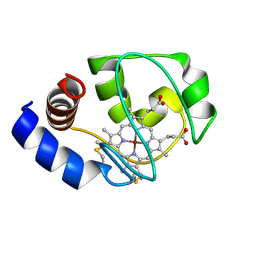 | | Solution Structure of Zinc-substituted cytochrome c | | Descriptor: | ZINC SUBSTITUTED HEME C, Zinc-substituted cytochrome c | | Authors: | Qian, C, Yao, Y, Tong, Y, Wang, J, Tang, W. | | Deposit date: | 2002-07-11 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of zinc-substituted cytochrome c.
J.Biol.Inorg.Chem., 8, 2003
|
|
1PCW
 
 | | Aquifex aeolicus KDO8PS in complex with cadmium and APP, a bisubstrate inhibitor | | Descriptor: | 1-DEOXY-6-O-PHOSPHONO-1-[(PHOSPHONOMETHYL)AMINO]-L-THREO-HEXITOL, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION | | Authors: | Xu, X, Wang, J, Grison, C, Petek, S, Coutrot, P, Birck, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2003-05-17 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of Novel Inhibitors of 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase.
Drug DES.DISCOVERY, 18, 2003
|
|
1NJ6
 
 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to alanine sulfamoyl adenylate | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2PQE
 
 | | Solution structure of proline-free mutant of staphylococcal nuclease | | Descriptor: | Thermonuclease | | Authors: | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
5U73
 
 | | Crystal structure of human Niemann-Pick C1 protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Niemann-Pick C1 protein, ... | | Authors: | Li, X, Wang, J, Blobel, G. | | Deposit date: | 2016-12-11 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7F4Y
 
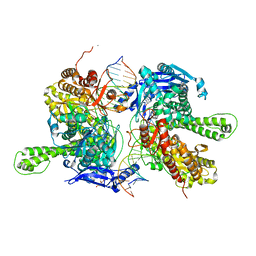 | | Crystal structure of replisomal dimer of DNA polymerase from bacteriophage RB69 with DNA duplexes | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*AP*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3'), ... | | Authors: | Youn, H.-S, Park, J, An, J.Y, Lee, Y, Eom, S.H, Wang, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of New Binary and Ternary DNA Polymerase Complexes From Bacteriophage RB69.
Front Mol Biosci, 8, 2021
|
|
6J6K
 
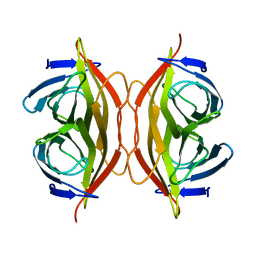 | | Apo-state streptavidin | | Descriptor: | Streptavidin | | Authors: | Fan, X, Wang, J, Lei, J.L, Wang, H.W. | | Deposit date: | 2019-01-15 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.
Nat Commun, 10, 2019
|
|
5MGW
 
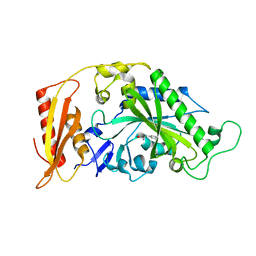 | | Kinetic and Structural Changes in HsmtPheRS, Induced by Pathogenic Mutations in Human FARS2 | | Descriptor: | PHENYLALANINE, Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
5MGU
 
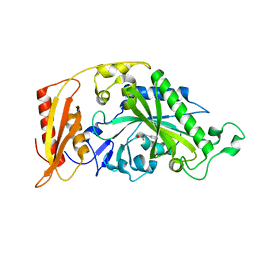 | | Kinetic and Structural Changes in HsmtPheRS, Induced by Pathogenic Mutations in Human FARS2 | | Descriptor: | PHENYLALANINE, Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
6J6J
 
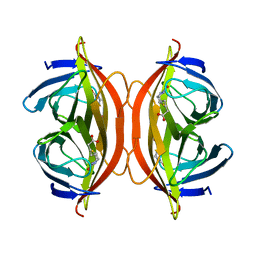 | | Biotin-bound streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Fan, X, Wang, J, Lei, J.L, Wang, H.W. | | Deposit date: | 2019-01-15 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.
Nat Commun, 10, 2019
|
|
5NEF
 
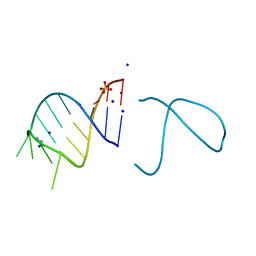 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with guanidine | | Descriptor: | GUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NDH
 
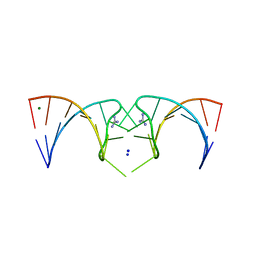 | | The structure of the G. violaceus guanidine II riboswitch P2 stem-loop | | Descriptor: | GUANIDINE, MAGNESIUM ION, RNA (5'-R(*GP*(CBV)P*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*GP*C)-3'), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
3P5N
 
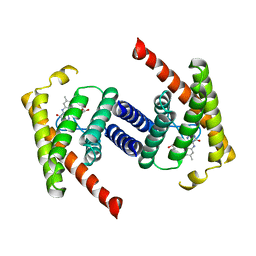 | |
5NEX
 
 | |
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57150865 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | Descriptor: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20017123 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65004492 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
3P49
 
 | | Crystal Structure of a Glycine Riboswitch from Fusobacterium nucleatum | | Descriptor: | GLYCINE, GLYCINE RIBOSWITCH, MAGNESIUM ION, ... | | Authors: | Butler, E.B, Wang, J, Xiong, Y, Strobel, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of cooperative ligand binding by the glycine riboswitch.
Chem.Biol., 18, 2011
|
|
