5XVB
 
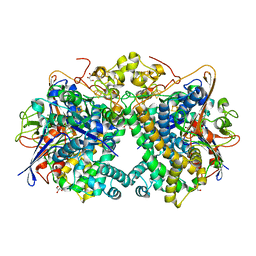 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
8HSB
 
 | |
1LD5
 
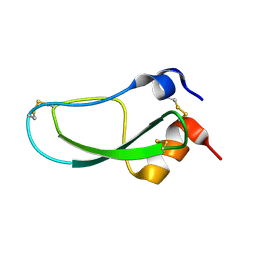 | | STRUCTURE OF BPTI MUTANT A16V | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR | | Authors: | Cierpicki, T, Otlewski, J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two variants of bovine pancreatic trypsin inhibitor (BPTI) reveal unexpected influence of mutations on protein structure and stability.
J.Mol.Biol., 321, 2002
|
|
6L84
 
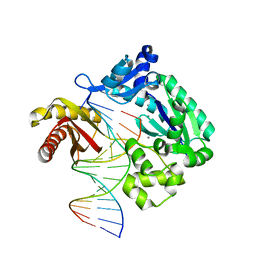 | | Complex of DNA polymerase IV and D-DNA duplex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(P*CP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Chung, H.S, An, J, Hwang, D. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The crystal structure of a natural DNA polymerase complexed with mirror DNA.
Chem.Commun.(Camb.), 56, 2020
|
|
8HIH
 
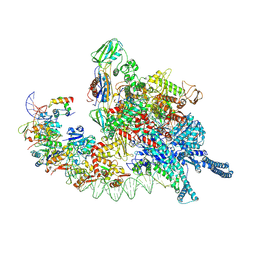 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2V8V
 
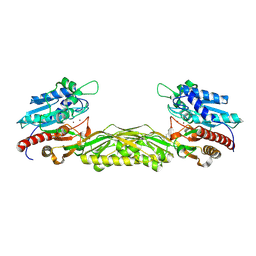 | | Crystal structure of mutant R322A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-ALANINE SYNTHASE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-15 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
8HMF
 
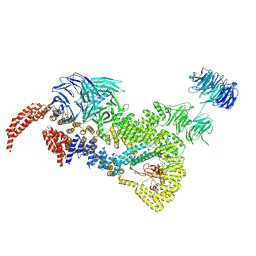 | | head module state 2 of Tetrahymena IFT-A | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transporter, WD40 repeat protein, ... | | Authors: | Ma, Y, Wu, J, Lei, M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
2V8H
 
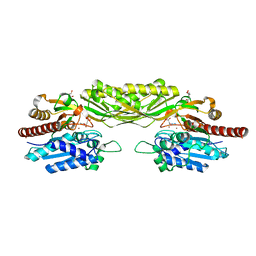 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri in complex with its substrate N-carbamyl-beta- alanine | | Descriptor: | BETA-ALANINE SYNTHASE, BICINE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
1U07
 
 | | Crystal Structure of the 92-residue C-term. part of TonB with significant structural changes compared to shorter fragments | | Descriptor: | TonB protein | | Authors: | Koedding, J, Killig, F, Polzer, P, Howard, S.P, Diederichs, K, Welte, W. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of a 92-residue c-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments
J.Biol.Chem., 280, 2005
|
|
4CRT
 
 | | Crystal structure of human monoamine oxidase B in complex with the multi-target inhibitor ASS234 | | Descriptor: | (E)-N-methyl-N-[[1-methyl-5-[3-[1-(phenylmethyl)piperidin-4-yl]propoxy]indol-2-yl]methyl]prop-1-en-1-amine, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Esteban, G, Allan, J, Samadi, A, Mattevi, A, Unzeta, M, Marco-Contelles, J, Binda, C, Ramsay, R.R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Analysis of the Irreversible Inhibition of Human Monoamine Oxidases by Ass234, a Multi-Target Compound Designed for Use in Alzheimer'S Disease.
Biochim.Biophys.Acta, 1844, 2014
|
|
1L8F
 
 | |
2V8D
 
 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-ALANINE SYNTHASE, ZINC ION | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
8HMD
 
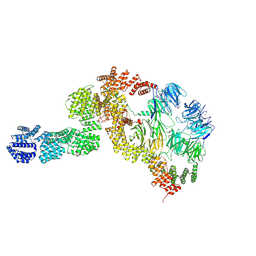 | | base module state 2 of Tetrahymena IFT-A | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 43 homolog, Tetratricopeptide repeat protein, ... | | Authors: | Ma, Y, Wu, J, Lei, M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
8B3T
 
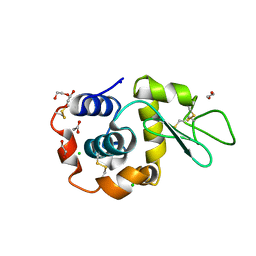 | | Hen Egg White Lysozyme 4s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
8B3U
 
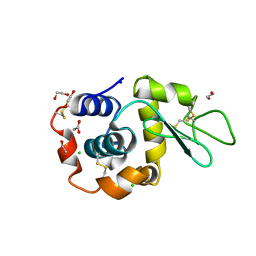 | | Hen Egg White Lysozyme 6s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
8B3V
 
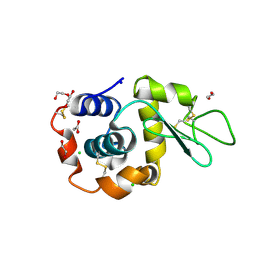 | | Hen Egg White Lysozyme 8s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
6LHV
 
 | | Structure of FANCA and FANCG Complex | | Descriptor: | Fanconi anemia complementation group A, Fanconi anemia complementation group G | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
2V62
 
 | | Structure of vaccinia-related kinase 2 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SERINE/THREONINE-PROTEIN KINASE VRK2, ... | | Authors: | Bunkoczi, G, Eswaran, J, Cooper, C, Fedorov, O, Keates, T, Rellos, P, Salah, E, Savitsky, P, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Pseudokinase Vrk3 Reveals a Degraded Catalytic Site, a Highly Conserved Kinase Fold, and a Putative Regulatory Binding Site.
Structure, 17, 2009
|
|
6TPK
 
 | | Crystal structure of the human oxytocin receptor | | Descriptor: | (3~{R},6~{R})-6-[(2~{S})-butan-2-yl]-3-(2,3-dihydro-1~{H}-inden-2-yl)-1-[(1~{R})-1-(2-methyl-1,3-oxazol-4-yl)-2-morpholin-4-yl-2-oxidanylidene-ethyl]piperazine-2,5-dione, CHOLESTEROL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waltenspuehl, Y, Schoeppe, J, Ehrenmann, J, Kummer, L, Plueckthun, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-08-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human oxytocin receptor.
Sci Adv, 6, 2020
|
|
8B3L
 
 | | Hen Egg White Lysozyme 2s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
6LPF
 
 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
8B1X
 
 | | Solution NMR structure of the single alpha helix peptide (P3-7)2 | | Descriptor: | P3-7_2 | | Authors: | Escobedo, A, Coles, M, Diercks, T, Garcia, J, Millet, O, Salvatella, X. | | Deposit date: | 2022-09-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A glutamine-based single alpha-helix scaffold to target globular proteins.
Nat Commun, 13, 2022
|
|
6LON
 
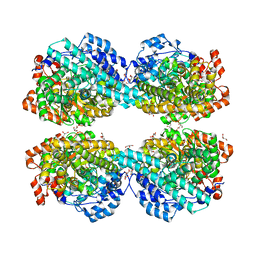 | | Crystal structure of HPSG | | Descriptor: | (2~{S})-2,3-bis(oxidanyl)propane-1-sulfonic acid, GLYCEROL, PFL2/glycerol dehydratase family glycyl radical enzyme, ... | | Authors: | Liu, J, Zhang, Y, Yuchi, Z. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two radical-dependent mechanisms for anaerobic degradation of the globally abundant organosulfur compound dihydroxypropanesulfonate.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LNK
 
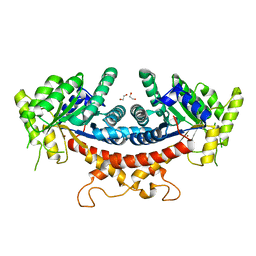 | | Candida albicans Fructose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Huang, Y, Cao, H, Ren, Y, Wan, J. | | Deposit date: | 2019-12-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
6M11
 
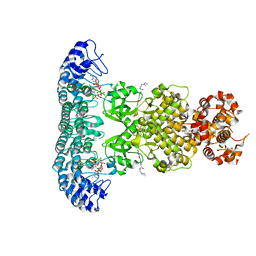 | | Crystal structure of Rnase L in complex with Sunitinib | | Descriptor: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, PHOSPHATE ION, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
