3LMF
 
 | | Crystal Structure of Nmul_A1745 protein from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR72 | | Descriptor: | Uncharacterized protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target NmR72
To be Published
|
|
4ZVI
 
 | | GYRASE B IN COMPLEX WITH 4,5-DIBROMOPYRROLAMIDE-BASED INHIBITOR | | Descriptor: | DNA gyrase subunit B, IODIDE ION, N-(4-{[(4,5-dibromo-1H-pyrrol-2-yl)carbonyl]amino}benzoyl)glycine | | Authors: | Zidar, N, Macut, H, Tomasic, T, Brvar, M, Montalvao, S, Tammela, P, Solmajer, T, Peterlin Masic, L, Ilas, J, Kikelj, D. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Phenyl-4,5-dibromopyrrolamides and N-Phenylindolamides as ATP Competitive DNA Gyrase B Inhibitors: Design, Synthesis, and Evaluation.
J.Med.Chem., 58, 2015
|
|
3LMM
 
 | | Crystal Structure of the DIP2311 protein from Corynebacterium diphtheriae, Northeast Structural Genomics Consortium Target CdR35 | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Uncharacterized protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Buchwald, W.A, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Northeast Structural Genomics Consortium Target CdR35
To be Published
|
|
5A0E
 
 | | Crystal structure of cyclophilin D in complex with CsA analogue, JW47. | | Descriptor: | JW47, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE F, MITOCHONDRIAL | | Authors: | Warne, J, Pryce, G, Hill, J, Shi, X, Lenneras, F, Puentes, F, Kip, M, Hilditch, L, Walker, P, Simone, M, Chan, A.W.E, Towers, G, Coker, A.R, Duchen, M, Szabadkai, G, Baker, D, Selwood, D.L. | | Deposit date: | 2015-04-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective Inhibition of the Mitochondrial Permeability Transition Pore Protects Against Neuro-Degeneration in Experimental Multiple Sclerosis.
J.Biol.Chem., 291, 2016
|
|
3LV4
 
 | | Crystal structure of the glycoside hydrolase, family 43 YxiA protein from Bacillus licheniformis. Northeast Structural Genomics Consortium Target BiR14. | | Descriptor: | ACETATE ION, CALCIUM ION, Glycoside hydrolase YxiA | | Authors: | Vorobiev, S, Abashidze, M, Seetharaman, J, Belote, R.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-19 | | Release date: | 2010-03-09 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of the glycoside hydrolase, family 43 YxiA protein from Bacillus licheniformis.
To be Published
|
|
4ZXK
 
 | |
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | Descriptor: | GLYCEROL, Putative aminotransferase | | Authors: | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
3LZ8
 
 | | Structure of a putative chaperone dnaj from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 2.9 a resolution. | | Descriptor: | Putative chaperone DnaJ | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Bearden, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Putative Chaperone Dnaj from Klebsiella Pneumoniae Subsp. Pneumoniae Mgh 78578 at 2.9 A Resolution.
To be Published
|
|
5A5A
 
 | | The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with PNP-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
3LZ9
 
 | | The Crystal Structure of 5-epi-aristolochene synthase M4 mutant complexed with (2-trans,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, MAGNESIUM ION | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
3LXM
 
 | | 2.00 Angstrom resolution crystal structure of a catalytic subunit of an aspartate carbamoyltransferase (pyrB) from Yersinia pestis CO92 | | Descriptor: | Aspartate carbamoyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of a catalytic subunit of an aspartate carbamoyltransferase (pyrB) from Yersinia pestis CO92
To be Published
|
|
3LUY
 
 | |
3LV8
 
 | | 1.8 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with TMP, thymidine-5'-diphosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-19 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with TMP, thymidine-5'-diphosphate and ADP
To be Published
|
|
5ADS
 
 | |
3LXX
 
 | | Crystal structure of human GTPase IMAP family member 4 | | Descriptor: | GTPase IMAP family member 4, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shen, Y, Nedyalkova, L, Tong, Y, Tempel, W, Mackenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Andrews, D.W, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human GTPase IMAP family member 4
to be published
|
|
3LYU
 
 | | Crystal Structure of the C-terminal domain (residues 83-215) of PF1911 hydrogenase from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR246A | | Descriptor: | Putative hydrogenase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target PfR246A
To be Published
|
|
3LYS
 
 | | Crystal Structure of the N-terminal domain of the Prophage pi2 protein 01 (integrase) from Lactococcus lactis, Northeast Structural Genomics Consortium Target KR124F | | Descriptor: | Prophage pi2 protein 01, integrase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-03-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Northeast Structural Genomics Consortium Target KR124F
To be Published
|
|
5A8F
 
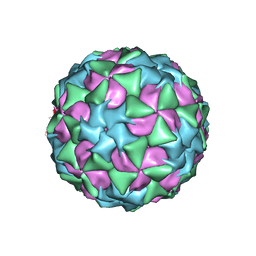 | | Structure and genome release mechanism of human cardiovirus Saffold virus-3 | | Descriptor: | GENOME POLYPHUMAN SAFFOLD VIRUS-3 VP3 PROTEIN, HUMAN SAFFOLD VIRUS-3 VP1, HUMAN SAFFOLD VIRUS-3 VP2 | | Authors: | Mullapudi, E, Novacek, J, Palkova, L, Kulich, P, Lindberg, M, vanKuppeveld, F.J.M, Plevka, P. | | Deposit date: | 2015-07-15 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and Genome Release Mechanism of Human Cardiovirus Saffold Virus-3.
J.Virol., 90, 2016
|
|
5A92
 
 | | 15K X-ray structure with Cefotaxime: Exploring the Mechanism of beta- Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5A4V
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A56
 
 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with 1-O-methyl-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights into Its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A7I
 
 | | Crystal structure of INPP5B in complex with biphenyl 3,3',4,4',5,5'- hexakisphosphate | | Descriptor: | Biphenyl 3,3',4,4',5,5'-hexakisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Mills, S.J, Silvander, C, Cozier, G, Potter, B.V.L, Norldund, P. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal Structures of Type-II Inositol Polyphosphate 5-Phosphatase Inpp5B with Synthetic Inositol Polyphosphate Surrogates Reveal New Mechanistic Insights for the Inositol 5-Phosphatase Family.
Biochemistry, 55, 2016
|
|
3M3Q
 
 | |
5A63
 
 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
5ABB
 
 | | Visualization of a polytopic membrane protein during SecY-mediated membrane insertion | | Descriptor: | GREEN-LIGHT ABSORBING PROTEORHODOPSIN, PROTEIN TRANSLOCASE SUBUNIT SECE, PROTEIN TRANSLOCASE SUBUNIT SECY | | Authors: | Bischoff, L, Wickles, S, Berninghausen, O, vanderSluis, E, Beckmann, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Visualization of a Polytopic Membrane Protein During Secy-Mediated Membrane Insertion.
Nat.Commun., 5, 2014
|
|
