8Q5P
 
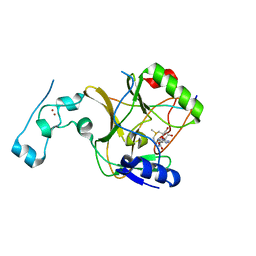 | | Structure of the lysine methyltransferase SETD2 in complex with a peptide derived from human tyrosine kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, Histone-lysine N-methyltransferase SETD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Mechaly, A, Le Coadou, L, Dupret, J.M, Haouz, A, Rodrigues Lima, F. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Structural and enzymatic evidence for the methylation of the ACK1 tyrosine kinase by the histone lysine methyltransferase SETD2.
Biochem.Biophys.Res.Commun., 695, 2024
|
|
4FX2
 
 | |
4MF0
 
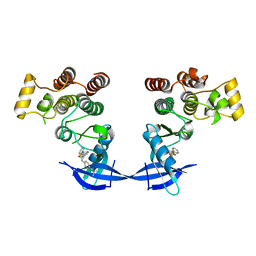 | | ITK kinase domain in complex with benzothiazole inhibitor compound 12a (1S,2S)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}-N-[6-(PYRIDIN-3-YL)-1,3-BENZOTHIAZOL-2-YL]CYCLOPROPANECARBOXAMIDE (12a) | | Descriptor: | (1S,2S)-2-{4-[(dimethylamino)methyl]phenyl}-N-[6-(pyridin-3-yl)-1,3-benzothiazol-2-yl]cyclopropanecarboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure-based design and synthesis of potent benzothiazole inhibitors of interleukin-2 inducible T cell kinase (ITK).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MF1
 
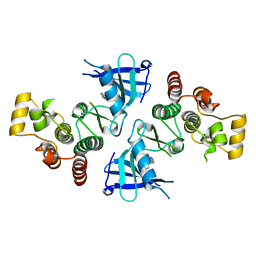 | | ITK kinase domain in complex with benzothiazole inhibitor 12b (1S,2S)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}-N-[6-(1H-PYRAZOL-4-YL)-1,3-BENZOTHIAZOL-2-YL]CYCLOPROPANECARBOXAMIDE | | Descriptor: | (1S,2S)-2-{4-[(dimethylamino)methyl]phenyl}-N-[6-(1H-pyrazol-4-yl)-1,3-benzothiazol-2-yl]cyclopropanecarboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Structure-based design and synthesis of potent benzothiazole inhibitors of interleukin-2 inducible T cell kinase (ITK).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7EYI
 
 | |
4MCY
 
 | | Immune Receptor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Citrullinated Vimentin, ... | | Authors: | Scally, S.W, Rossjohn, J. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis.
J.Exp.Med., 210, 2013
|
|
4MKW
 
 | |
4MRB
 
 | | Wild Type Human Transthyretin pH 7.5 | | Descriptor: | CALCIUM ION, Transthyretin | | Authors: | Chen, W.J, Wood, S.P. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7JQ8
 
 | | Solution NMR structure of human Brd3 ET domain | | Descriptor: | Bromodomain-containing protein 3, Integrase peptide | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, J.M, Montelione, G.T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JMY
 
 | |
7JYN
 
 | | Solution NMR structure of human Brd3 ET complexed with NSD3(148-184) peptide | | Descriptor: | Bromodomain-containing protein 3, Histone-lysine N-methyltransferase NSD3 | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7KJP
 
 | | Disulfide Stabilized Norovirus GI.1 VLP Shell Region | | Descriptor: | Capsid protein VP1 | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Disulfide stabilization of human norovirus GI.1 virus-like particles focuses immune response toward blockade epitopes.
NPJ Vaccines, 5, 2020
|
|
7VO8
 
 | | Structure of AtHPPD complexed with LSY-1 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, N-[6-[[9-(dimethylamino)-12H-benzo[a]phenoxazin-5-yl]amino]hexyl]-4-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-[oxidanyl(oxidanylidene)-$l^4-azanyl]benzamide | | Authors: | Yang, G.-F, Dong, J. | | Deposit date: | 2021-10-12 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In vivo diagnostics of abiotic plant stress responses via in situ real-time fluorescence imaging
Plant Physiol., 190, 2022
|
|
7X12
 
 | | Crystal structure of ME1 in complex with NADPH | | Descriptor: | MANGANESE (II) ION, NADP-dependent malic enzyme, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and Characterization of a Novel Allosteric Small-Molecule Inhibitor of NADP + -Dependent Malic Enzyme 1.
Biochemistry, 61, 2022
|
|
7X11
 
 | | Crystal structure of ME1 in complex with NADPH | | Descriptor: | 6-[(7-methyl-2-propyl-imidazo[4,5-b]pyridin-4-yl)methyl]-2-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]-1,3-benzothiazole, MANGANESE (II) ION, NADP-dependent malic enzyme, ... | | Authors: | Amano, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and Characterization of a Novel Allosteric Small-Molecule Inhibitor of NADP + -Dependent Malic Enzyme 1.
Biochemistry, 61, 2022
|
|
7VLI
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2220 | | Descriptor: | 1-[(3~{S},6~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VLG
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2201 | | Descriptor: | 1-[(5~{R},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-5-propan-2-yl-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VLH
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2219 | | Descriptor: | 1-[(3~{S},6~{R},18~{R})-3,6-bis(4-azanylbutyl)-2,5,8,11,14,17-hexakis(oxidanylidene)-1,4,7,10,13,16-hexazacyclodocos-18-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7Z8O
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-03-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YZY
 
 | | pMMO structure from native membranes by cryoET and STA | | Descriptor: | Methane monooxygenase subunit C2, Particulate methane monooxygenase alpha subunit, Particulate methane monooxygenase beta subunit | | Authors: | Zhu, Y, Ni, T, Zhang, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and activity of particulate methane monooxygenase arrays in methanotrophs.
Nat Commun, 13, 2022
|
|
8CKW
 
 | |
8CL4
 
 | |
8CL1
 
 | |
8CKZ
 
 | |
8CKX
 
 | |
