6LG0
 
 | | Crystal structure of SbCGTa in complex with UDP | | Descriptor: | SbCGTa, URIDINE-5'-DIPHOSPHATE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KUU
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
8YMO
 
 | | OSCA1.1-F516A pre-open 1 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
8YMP
 
 | | OSCA1.1-F516A nanodisc in LPC | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
8YMQ
 
 | | OSCA1.1-F516A nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
8YMM
 
 | | OSCA1.1-F516A open | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
8YMN
 
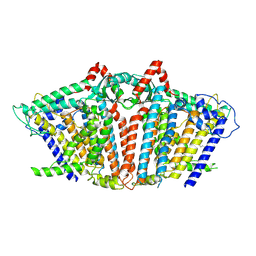 | | OSCA1.1-F516A pre-open 2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Protein OSCA1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-03-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels.
Nat Commun, 15, 2024
|
|
5XN4
 
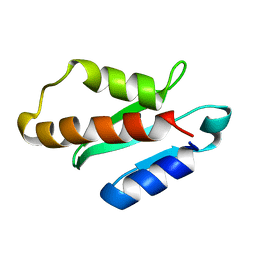 | | Anti-CRISPR protein AcrIIA4 | | Descriptor: | Anti-CRISPR AcrIIA4 | | Authors: | Suh, J.-Y, Kim, I. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor.
Sci Rep, 8, 2018
|
|
8ZAM
 
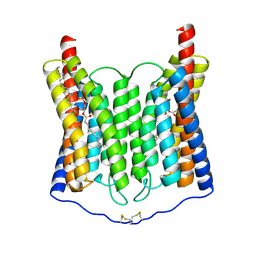 | | EndoChR2 channelrhodopsin | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Channelrhodopsins with distinct chromophores and binding patterns.
Nat Commun, 15, 2024
|
|
8ZAO
 
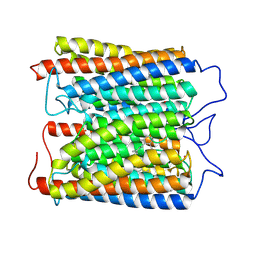 | | ExoKCR1 channelrhodopsin | | Descriptor: | POTASSIUM ION, RETINAL, exoKCR1 | | Authors: | Zhang, M.F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Channelrhodopsins with distinct chromophores and binding patterns.
Nat Commun, 15, 2024
|
|
8ZAQ
 
 | | ExoC110T class 2 channelrhodopsin | | Descriptor: | C110T class2, POTASSIUM ION, RETINAL | | Authors: | Zhang, M.F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Channelrhodopsins with distinct chromophores and binding patterns.
Nat Commun, 15, 2024
|
|
8ZAN
 
 | | ExoChR2 channelrhodopsin | | Descriptor: | Archaeal-type opsin 2, RETINAL | | Authors: | Zhang, M.F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Channelrhodopsins with distinct chromophores and binding patterns.
Nat Commun, 15, 2024
|
|
8ZAP
 
 | | ExoC110T class 1 channelrhodopsin | | Descriptor: | C110T-class1, POTASSIUM ION, RETINAL | | Authors: | Zhang, M.F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Channelrhodopsins with distinct chromophores and binding patterns.
Nat Commun, 15, 2024
|
|
7O0N
 
 | |
4UT3
 
 | | X-ray structure of the human PP1 gamma catalytic subunit treated with hydrogen peroxide | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Zeh Silva, M, Kopec, J, Fotinou, D, Steiner, R.A. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeted Redox Inhibition of Protein Phosphatase 1 by Nox4 Regulates Eif2Alpha-Mediated Stress Signaling.
Embo J., 35, 2016
|
|
4UT2
 
 | | X-ray structure of the human PP1 gamma catalytic subunit treated with ascorbate | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Kopec, J, Zeh Silva, M, Fotinou, C, Steiner, R.A. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeted Redox Inhibition of Protein Phosphatase 1 by Nox4 Regulates Eif2Alpha-Mediated Stress Signaling.
Embo J., 35, 2016
|
|
3O71
 
 | | Crystal structure of ERK2/DCC peptide complex | | Descriptor: | Mitogen-activated protein kinase 1, Peptide of Deleted in Colorectal Cancer, THIOCYANATE ION | | Authors: | Ma, W.F, Shang, Y, Wei, Z.Y, Wen, W.Y, Wang, W.N, Zhang, M.J. | | Deposit date: | 2010-07-30 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphorylation of DCC by ERK2 is facilitated by direct docking of the receptor P1 domain to the kinase
Structure, 18, 2010
|
|
6L5K
 
 | | ARF5 Aux/IAA17 Complex | | Descriptor: | Auxin response factor 5, Auxin-responsive protein IAA17 | | Authors: | Ryu, K.S, Suh, J.Y, Cha, S.Y, Kim, Y.I, Park, C.K. | | Deposit date: | 2019-10-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Determinants of PB1 Domain Interactions in Auxin Response Factor ARF5 and Repressor IAA17.
J.Mol.Biol., 432, 2020
|
|
8BHY
 
 | |
8BH3
 
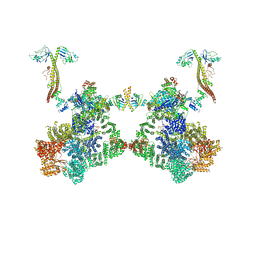 | | DNA-PK Ku80 mediated dimer bound to PAXX | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Hardwick, S.W, Chaplin, A.K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
8BHV
 
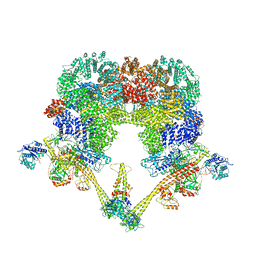 | | DNA-PK XLF mediated dimer bound to PAXX | | Descriptor: | DNA (24-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Hardwick, S.W, Chaplin, A.K. | | Deposit date: | 2022-11-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
4UXD
 
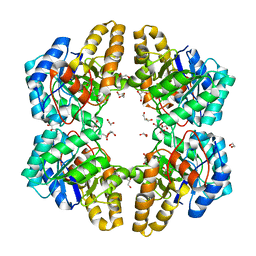 | | 2-keto 3-deoxygluconate aldolase from Picrophilus torridus | | Descriptor: | 1,2-ETHANEDIOL, 2-DEHYDRO-3-DEOXY-D-GLUCONATE/2-DEHYDRO-3-DEOXY-PHOSPHOGLUCONATE ALDOLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priftis, A, Zaitsev, V, Reher, M, Johnsen, U, Danson, M.J, Taylor, G.L, Schoenheit, P, Crennell, S.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
7O49
 
 | |
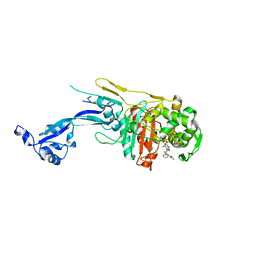
7O4A
 
 | |
7O4C
 
 | |
