2WS7
 
 | | Semi-synthetic analogue of human insulin ProB26-DTI | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WTJ
 
 | | CRYSTAL STRUCTURE OF CHK2 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5-(2,3-DIHYDROTHIENO[3,4-B][1,4]DIOXIN-5-YL)-N-[2-(DIMETHYLAMINO)ETHYL]PYRIDINE-3-CARBOXAMIDE, CHECKPOINT KINASE 2, ... | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-16 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
6XZR
 
 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 1 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6Y16
 
 | | CRYSTAL STRUCTURE OF TMARGBP DOMAIN 1 IN COMPLEX WITH THE GUANIDINIUM ION | | Descriptor: | 1,2-ETHANEDIOL, Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, ... | | Authors: | Ruggiero, A, Balasco, N, Smaldone, G, Graziano, G, Vitagliano, L. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium binding to proteins: The intriguing effects on the D1 and D2 domains of Thermotoga maritima Arginine Binding Protein and a comprehensive analysis of the Protein Data Bank.
Int.J.Biol.Macromol., 163, 2020
|
|
2WRS
 
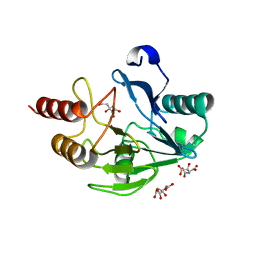 | | Crystal Structure of the Mono-Zinc Metallo-beta-lactamase VIM-4 from Pseudomonas aeruginosa | | Descriptor: | BETA-LACTAMASE VIM-4, CITRATE ANION, CITRIC ACID, ... | | Authors: | Lassaux, P, Hamel, M, Gulea, M, Delbruck, H, Traore, D.A.K, Mercuri, P.S, Horsfall, L, Dehareng, D, Gaumont, A.-C, Frere, J.-M, Ferrer, J.-L, Hoffmann, K, Galleni, M, Bebrone, C. | | Deposit date: | 2009-09-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-Beta-Lactamases.
J.Med.Chem., 53, 2010
|
|
2WVE
 
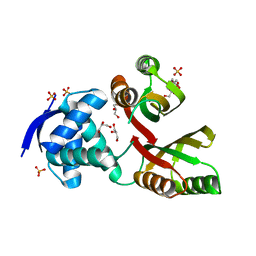 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, GLYCEROL, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
2Z5F
 
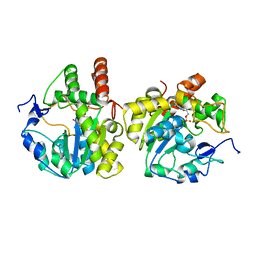 | | Human sulfotransferase Sult1b1 in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase family cytosolic 1B member 1 | | Authors: | Dombrovski, L, Pegasova, T, Wu, H, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-07 | | Release date: | 2007-08-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human sulfotransferases SULT1B1 and SULT1C1 complexed with the cofactor product adenosine-3'-5'-diphosphate (PAP)
Proteins, 64, 2006
|
|
2X7V
 
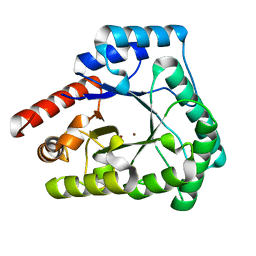 | | Crystal structure of Thermotoga maritima endonuclease IV in the presence of zinc | | Descriptor: | PROBABLE ENDONUCLEASE 4, ZINC ION | | Authors: | Tomanicek, S.J, Hughes, R.C, Ng, J.D, Coates, L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Endonuclease Iv Homologue from Thermotoga Maritima in the Presence of Active-Site Divalent Metal Ions
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2X7Z
 
 | | Crystal Structure of the SAP97 PDZ2 I342W C378A mutant protein domain | | Descriptor: | AMMONIUM ION, DISKS LARGE HOMOLOG 1, IMIDAZOLE | | Authors: | Haq, S.R, Jurgens, M.C, Chi, C.N, Elfstrom, L, Koh, C.S, Selmer, M, Gianni, S, Jemth, P. | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Plastic Energy Landscape of Protein Folding: A Triangular Folding Mechanism with an Equilibrium Intermediate for a Small Protein Domain.
J.Biol.Chem., 285, 2010
|
|
2WSP
 
 | | Thermotoga maritima alpha-L-fucosynthase, TmD224G, in complex with alpha-L-Fuc-(1-2)-beta-L-Fuc-N3 | | Descriptor: | ALPHA-L-FUCOSIDASE, PUTATIVE, alpha-L-fucopyranose-(1-2)-beta-L-fucosyl-azide | | Authors: | Sulzenbacher, G, Lipski, A, Cobucci-Ponzano, B, Conte, F, Bedini, E, Corsaro, M.M, Parrilli, M, Dal Piaz, F, Lepore, L, Rossi, M, Moracci, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Beta-Glycosyl Azides as Substrates for Alpha-Glycosynthases: Preparation of Novel Efficient Alpha-L-Fucosynthases
Chem.Biol., 16, 2009
|
|
6YDD
 
 | | X-ray structure of LPMO. | | Descriptor: | COPPER (II) ION, LPMO lytic polysaccharide monooxygenase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tandrup, T, Tryfona, T, Frandsen, K.E.H, Johansen, K.S, Dupree, P, Lo Leggio, L. | | Deposit date: | 2020-03-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Oligosaccharide Binding and Thermostability of Two Related AA9 Lytic Polysaccharide Monooxygenases.
Biochemistry, 59, 2020
|
|
6YL5
 
 | | Crystal structure of the SAM-SAH riboswitch with SAH | | Descriptor: | Chains: A,B,C,D,E,F,G,H,I,J,K,L, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YLH
 
 | |
6YMI
 
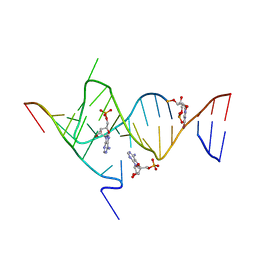 | | Crystal structure of the SAM-SAH riboswitch with AMP. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE MONOPHOSPHATE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YS3
 
 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
6Y2X
 
 | | RING-DTC domains of Deltex 2, Form 2 | | Descriptor: | Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination.
Sci Adv, 6, 2020
|
|
2UV7
 
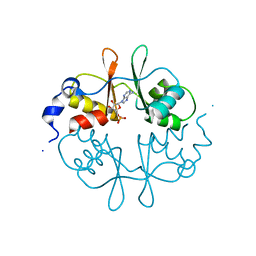 | | Crystal Structure of a CBS domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ADENOSINE MONOPHOSPHATE | | Authors: | Day, P, Sharff, A, Parra, L, Cleasby, A, Williams, M, Horer, S, Nar, H, Redemann, N, Tickle, I, Yon, J. | | Deposit date: | 2007-03-09 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Cbs-Domain Pair from the Regulatory Gamma1 Subunit of Human Ampk in Complex with AMP and Zmp.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6Y72
 
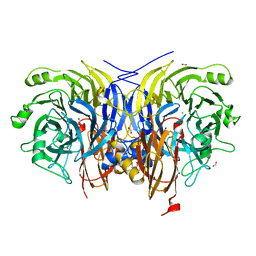 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H178A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y3J
 
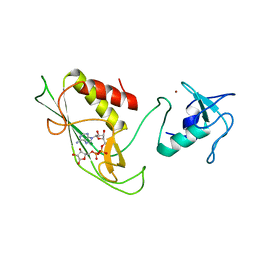 | | RING-DTC domains of Deltex 2, bound to ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Gabrielssen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination.
Sci Adv, 6, 2020
|
|
6Y8K
 
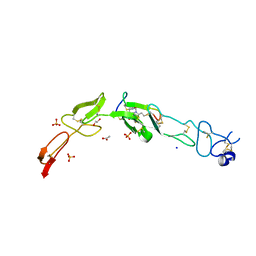 | | Crystal structure of CD137 in complex with the cyclic peptide BCY10916 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Upadhyaya, P, Kublin, J, Dods, R, Kristensson, J, Lahdenranta, J, Kleyman, M, Repash, E, Ma, J, Mudd, G, Van Rietschoten, K, Haines, E, Harrison, H, Beswick, P, Chen, L, McDonnell, K, Battula, S, Hurov, K, Keen, N. | | Deposit date: | 2020-03-05 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Anticancer immunity induced by a synthetic tumor-targeted CD137 agonist.
J Immunother Cancer, 9, 2021
|
|
2ZBV
 
 | | Crystal structure of uncharacterized conserved protein from Thermotoga maritima | | Descriptor: | ADENOSINE, Uncharacterized conserved protein | | Authors: | Ebihara, A, Fujimoto, Y, Kagawa, W, Fujikawa, N, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Thermotoga maritima
To be Published
|
|
2VL7
 
 | | Structure of S. tokodaii Xpd4 | | Descriptor: | PHOSPHATE ION, XPD | | Authors: | Naismith, J.H, Johnson, K.A, Oke, M, McMahon, S.A, Liu, L, White, M.F, Zawadski, M, Carter, L.G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the DNA Repair Helicase Xpd.
Cell(Cambridge,Mass.), 133, 2008
|
|
2VBL
 
 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*DA*DA*DA*DA*DG*DG*DC*DA*DG*DAP)-3', 5'-D(*DA*DG*DG*DA*DT*DC*DC*DT*DA*DAP)-3', 5'-D(*DT*DC*DT*DG*DC*DC*DT*DT*DT*DT*DT*DT *DGP*DAP)-3', ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
6Y9T
 
 | | Family GH13_31 enzyme | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Andersen, S, Poulsen, J.C.N, Moeller, M.S, Abou Hachem, M, Lo Leggio, L. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | An 1,4-alpha-Glucosyltransferase Defines a New Maltodextrin Catabolism Scheme in Lactobacillus acidophilus.
Appl.Environ.Microbiol., 86, 2020
|
|
6Y5P
 
 | | RING-DTC domain of Deltex1 bound to NAD | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase DTX1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases.
Sci Adv, 6, 2020
|
|
