6DBM
 
 | | Tyk2 with compound 23 | | Descriptor: | Non-receptor tyrosine-protein kinase TYK2, [(1S)-2,2-difluorocyclopropyl][(1R,5S)-3-{2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
6DBN
 
 | | Jak1 with compound 23 | | Descriptor: | Tyrosine-protein kinase JAK1, [(1S)-2,2-difluorocyclopropyl][(1R,5S)-3-{2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
6G5J
 
 | | Secreted phospholipase A2 type X in complex with ligand | | Descriptor: | (3~{R})-3-[3-[2-aminocarbonyl-6-(trifluoromethyloxy)indol-1-yl]phenyl]butanoic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Selective sPLA2-X Inhibitor (-)-2-{2-[Carbamoyl-6-(trifluoromethoxy)-1H-indol-1-yl]pyridine-2-yl}propanoic Acid.
ACS Med Chem Lett, 9, 2018
|
|
5JO3
 
 | | PDE5A for NaV1.7 | | Descriptor: | 1-(5-chloro-6-methoxypyridin-3-yl)-3-methyl-N-(methylsulfonyl)-1H-indazole-5-carboxamide, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Storer, I, Chrencik, J. | | Deposit date: | 2016-05-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PDE5A for NaV1.7
To be published
|
|
5GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, GLUTATHIONYLSPERMIDINE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, GLUTATHIONYLSPERMIDINE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
3DY7
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | (5S)-4,5-difluoro-6-[(2-fluoro-4-iodophenyl)imino]-N-(2-hydroxyethoxy)cyclohexa-1,3-diene-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-07-25 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Beyond the MEK-pocket: can current MEK kinase inhibitors be utilized to synthesize novel type III NCKIs? Does the MEK-pocket exist in kinases other than MEK?
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4FF8
 
 | | Inhibitor bound structure of the kinase domain of the murine receptor tyrosine kinase TYRO3 (Sky) | | Descriptor: | 4-(cyclopentylamino)-2-[(2-methoxybenzyl)amino]-N-[3-(2-oxopyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Tyrosine-protein kinase receptor TYRO3 | | Authors: | Ohren, J.F, Powell, N.A, Kohrt, J, Perrin, L.A. | | Deposit date: | 2012-05-31 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly selective 2,4-diaminopyrimidine-5-carboxamide inhibitors of Sky kinase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7P2D
 
 | | Structure of alphaMbeta2/Cd11bCD18 headpiece in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2021-07-05 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the function-modulating effects of nanobody binding to the integrin receptor alpha M beta 2.
J.Biol.Chem., 298, 2022
|
|
6EUC
 
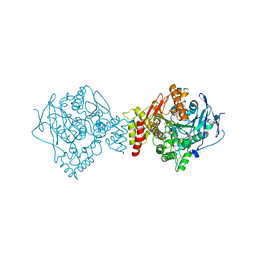 | | Reactivating oxime bound to Tc AChE's catalytic gorge. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-10-30 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.21998858 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
5C9C
 
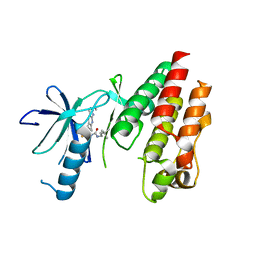 | | CRYSTAL STRUCTURE OF BRAF(V600E) IN COMPLEX WITH LY3009120 COMPND | | Descriptor: | 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Edwards, T, Abendroth, J, Chun, L. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers.
Cancer Cell, 28, 2015
|
|
4FEQ
 
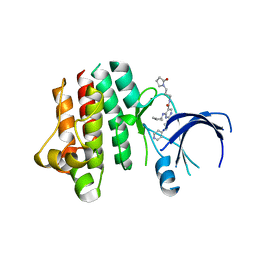 | | Inhibitor bound structure of the kinase domain of the murine receptor tyrosine kinase TYRO3 (Sky) | | Descriptor: | 4-(cyclopentylamino)-N-[3-(2-oxopyrrolidin-1-yl)propyl]-2-{[2-(pyridin-4-yl)ethyl]amino}pyrimidine-5-carboxamide, Tyrosine-protein kinase receptor TYRO3 | | Authors: | Ohren, J.F, Powell, N.A, Kohrt, J.T, Perrin, L.A. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Highly selective 2,4-diaminopyrimidine-5-carboxamide inhibitors of Sky kinase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6EWK
 
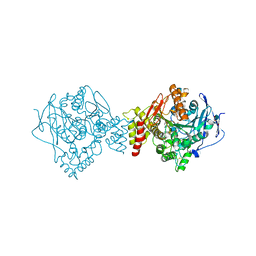 | | T. californica AChE in complex with a 3-hydroxy-2-pyridine aldoxime. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
7PV1
 
 | |
7PUZ
 
 | |
7PV0
 
 | | Crystal structure of a Mic60-Mic19 fusion protein | | Descriptor: | MICOS complex subunit MIC60,MICOS complex subunit MIC60-MIC19,Mic60-Mic19, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Funck, K, Bock-Bierbaum, T, Daumke, O. | | Deposit date: | 2021-10-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into crista junction formation by the Mic60-Mic19 complex.
Sci Adv, 8, 2022
|
|
8T9G
 
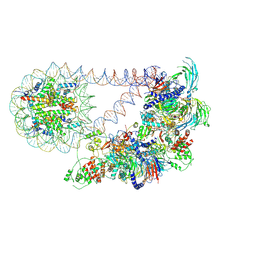 | | Automethylated PRC2 dimer bound to nucleosome | | Descriptor: | DNA (226-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Sauer, P.V, Pavlenko, E, Nogales, E, Poepsel, S. | | Deposit date: | 2023-06-23 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Activation of automethylated PRC2 by dimerization on chromatin.
Mol.Cell, 2024
|
|
8TB9
 
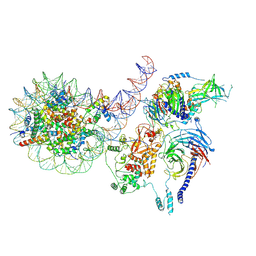 | | PRC2-J119-450 monomer bound to H1-nucleosome | | Descriptor: | DNA (226-MER), Histone H1.0, Histone H2A type 1, ... | | Authors: | Sauer, P.V, Cookis, T, Pavlenko, E, Nogales, E, Poepsel, S. | | Deposit date: | 2023-06-28 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of automethylated PRC2 by dimerization on chromatin.
Mol.Cell, 2024
|
|
8TAS
 
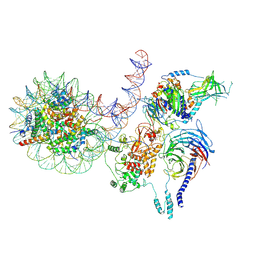 | | PRC2 monomer bound to nucleosome | | Descriptor: | DNA (226-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Sauer, P.V, Pavlenko, E, Nogales, E, Poepsel, S. | | Deposit date: | 2023-06-27 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Activation of automethylated PRC2 by dimerization on chromatin.
Mol.Cell, 2024
|
|
1BF3
 
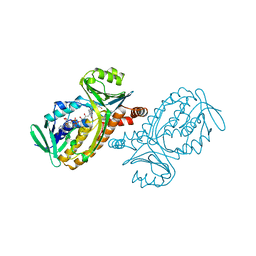 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND ARG 42 REPLACED BY LYS (R42K), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lys42 and Ser42 variants of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens reveal that Arg42 is essential for NADPH binding.
Eur.J.Biochem., 253, 1998
|
|
1BGJ
 
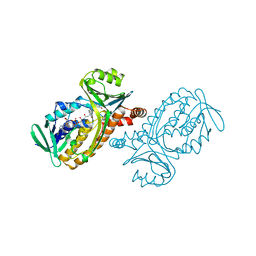 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND HIS 162 REPLACED BY ARG (H162R), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
J.Biol.Chem., 273, 1998
|
|
1BKW
 
 | | p-Hydroxybenzoate hydroxylase (phbh) mutant with cys116 replaced by ser (c116s) and arg44 replaced by lys (r44k), in complex with fad and 4-hydroxybenzoic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H, Schreuder, H.A, Van Berkel, W.J. | | Deposit date: | 1998-07-13 | | Release date: | 1998-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of mutant Arg44Lys of 4-hydroxybenzoate hydroxylase implications for NADPH binding.
Eur.J.Biochem., 231, 1995
|
|
7ZO0
 
 | | Crystal structure of catalytic inactive unliganded form of FucOB, a GH95 family alpha-1,2-fucosidase from Akkermansia muciniphila | | Descriptor: | GH95 family alpha-1,2-fucosidase, GLYCEROL | | Authors: | Anso, I, Cifuente, J.O, Trastoy, B, Guerin, M.E. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Turning universal O into rare Bombay type blood.
Nat Commun, 14, 2023
|
|
1PBB
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2,4-DIHYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
7NP9
 
 | |
1PBC
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
