2CI0
 
 | | High throughput screening and x-ray crystallography assisted evaluation of small molecule scaffolds for CYP51 inhibitors | | Descriptor: | (2R)-2-PHENYL-N-PYRIDIN-4-YLBUTANAMIDE, CYTOCHROME P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M, Kim, Y, Yermalitskaya, L.V, Von Kries, J.P, Waterman, M.R. | | Deposit date: | 2006-03-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Small Molecule Scaffolds for Cyp51 Inhibitors Identified by High Throughput Screening and Defined by X-Ray Crystallography
Antimicrob.Agents Chemother., 51, 2007
|
|
2CA0
 
 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Yc-17-Bound Cytochrome P450 Pikc (Cyp107L1)
To be Published
|
|
2C7X
 
 | | Crystal structure of narbomycin-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | CYTOCHROME P450 MONOOXYGENASE, NARBOMYCIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-29 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2CD8
 
 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | Deposit date: | 2006-01-20 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2B2T
 
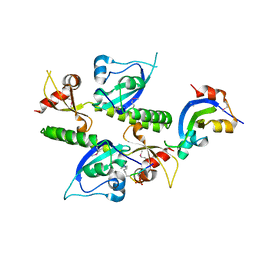 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and phosphothreonine 3 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 tail | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
3ZGX
 
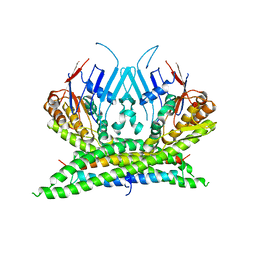 | | Crystal structure of the kleisin-N SMC interface in prokaryotic condensin | | Descriptor: | CHROMOSOME PARTITION PROTEIN SMC, SEGREGATION AND CONDENSATION PROTEIN A | | Authors: | Burmann, F, Shin, H, Basquin, J, Soh, Y, Gimenez, V, Kim, Y, Oh, B, Gruber, S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Asymmetric Smc-Kleisin Bridge in Prokaryotic Condensin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2B2Y
 
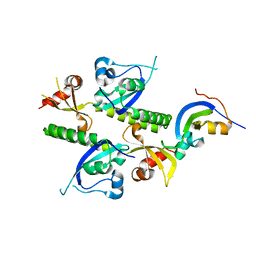 | | Tandem chromodomains of human CHD1 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2U
 
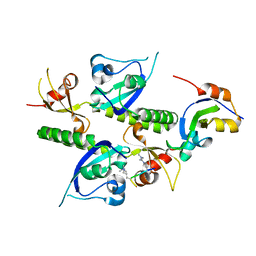 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2V
 
 | | Crystal structure analysis of human CHD1 chromodomains 1 and 2 bound to histone H3 resi 1-15 MeK4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
1L1S
 
 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
1XG8
 
 | | Crystal Structure of Protein of Unknown Function SA0789 from Staphylococcus aureus | | Descriptor: | hypothetical protein SA0798 | | Authors: | Rotella, F.J, Zhang, R.G, Kim, Y, Quartey, P, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A crystal structure of hypothetical protein SA0798 from Staphylococcus aureus
To be Published
|
|
4FK1
 
 | | Crystal Structure of Putative Thioredoxin Reductase TrxB from Bacillus anthracis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Crystal Structure of Putative Thioredoxin Reductase TrxB from Bacillus anthracis
To be Published
|
|
1GWI
 
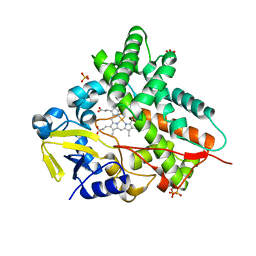 | | The 1.92 A structure of Streptomyces coelicolor A3(2) CYP154C1: A new monooxygenase that functionalizes macrolide ring systems | | Descriptor: | CYTOCHROME P450 154C1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Podust, L.M, Kim, Y, Arase, M, Neely, B.A, Beck, B.J, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2002-03-15 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.92 A Structure of Streptomyces Coelicolor A3(2) Cyp154C1: A New Monooxygenase that Functionalizes Macrolide Ring Systems
J.Biol.Chem., 278, 2003
|
|
1NRW
 
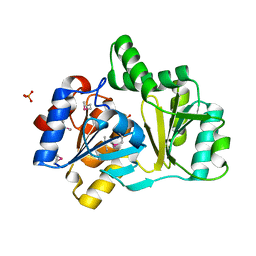 | | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS | | Descriptor: | CALCIUM ION, PHOSPHATE ION, hypothetical protein, ... | | Authors: | Cuff, M.E, Kim, Y, Zhang, R, Joachimiak, A, Collart, F, Quartey, P, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-25 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS
To be Published
|
|
1MT6
 
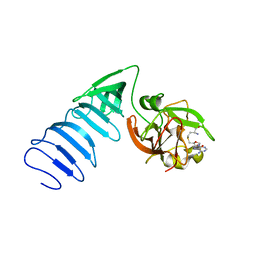 | | Structure of histone H3 K4-specific methyltransferase SET7/9 with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET9 | | Authors: | Jacobs, S.A, Harp, J.M, Devarakonda, S, Kim, Y, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active site of the SET domain is constructed on a knot
Nat.Struct.Biol., 9, 2002
|
|
1ODO
 
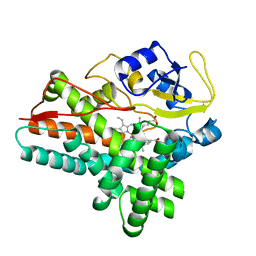 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | Authors: | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
4GUD
 
 | | Crystal Structure of Amidotransferase HisH from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Crystal Structure of Amidotransferase HisH from Vibrio cholerae.
To be Published
|
|
1K3R
 
 | | Crystal Structure of the Methyltransferase with a Knot from Methanobacterium thermoautotrophicum | | Descriptor: | conserved protein MT0001 | | Authors: | Zarembinski, T.I, Kim, Y, Peterson, K, Christendat, D, Dharamsi, A, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-03 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deep trefoil knot implicated in RNA binding found in an archaebacterial protein.
Proteins, 50, 2003
|
|
1MUF
 
 | | Structure of histone H3 K4-specific methyltransferase SET7/9 | | Descriptor: | SET9 | | Authors: | Jacobs, S.A, Harp, J.M, Devarakonda, S, Kim, Y, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The active site of the SET domain is constructed on a knot
Nat.Struct.Biol., 9, 2002
|
|
2N50
 
 | | Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis | | Descriptor: | Acyl carrier protein | | Authors: | Park, Y, Jung, M, Song, H, Jeong, K, Kim, Y. | | Deposit date: | 2015-07-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis.
J. Biol. Chem., 291, 2016
|
|
1NNI
 
 | | Azobenzene Reductase from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein yhda | | Authors: | Cuff, M.E, Kim, Y, Maj, L, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-13 | | Release date: | 2003-07-29 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azobenzene Reductase from Bacillus subtilis
To be Published, 2003
|
|
4IQR
 
 | | Multi-Domain Organization of the HNF4alpha Nuclear Receptor Complex on DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), Hepatocyte nuclear factor 4-alpha, ... | | Authors: | Chandra, V, Huang, P, Kim, Y, Rastinejad, F. | | Deposit date: | 2013-01-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Multidomain integration in the structure of the HNF-4 alpha nuclear receptor complex.
Nature, 495, 2013
|
|
4IR0
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
2HLY
 
 | | The crystal structure of genomics APC5867 | | Descriptor: | Hypothetical protein Atu2299 | | Authors: | Dong, A, Xu, X, Zheng, H, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-10 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of genomics APC5867
TO BE PUBLISHED
|
|
2FBQ
 
 | | The crystal structure of transcriptional regulator PA3006 | | Descriptor: | probable transcriptional regulator | | Authors: | Lunin, V.V, Skarina, T, Onopriyenko, O, Kim, Y, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transcriptional regulator PA3006
To be Published
|
|
