7SN0
 
 | | Crystal structure of spike protein receptor binding domain of escape mutant SARS-CoV-2 from immunocompromised patient (d146*) in complex with human receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Clark, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN1
 
 | |
7SN3
 
 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN2
 
 | | Structure of human SARS-CoV-2 neutralizing antibody C1C-A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Yang, P, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7TCL
 
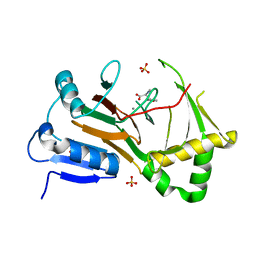 | | Crystal structure of P.IsnB complexed with tyrosine isonitrile | | Descriptor: | (2S)-3-(4-hydroxyphenyl)-2-isocyanopropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-12-26 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Elucidation of divergent desaturation pathways in the formation of vinyl isonitrile and isocyanoacrylate.
Nat Commun, 13, 2022
|
|
8UXS
 
 | |
6IRR
 
 | | Solution structure of DISC1/ATF4 complex | | Descriptor: | Disrupted in schizophrenia 1 homolog,Cyclic AMP-dependent transcription factor ATF-4 | | Authors: | Ye, F, Yu, C, Zhang, M. | | Deposit date: | 2018-11-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.
Mol. Psychiatry, 2019
|
|
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
5XK5
 
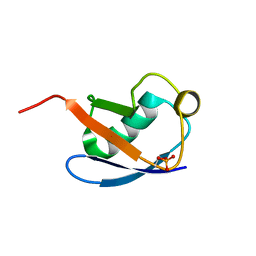 | | Relaxed state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Xu, D, Zhou, G, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
