9BWX
 
 | | Consensus full-complex model for preturnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
6JBJ
 
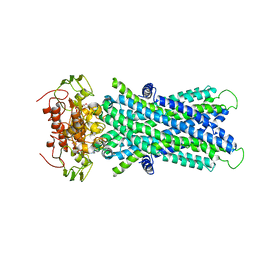 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | Authors: | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
7URG
 
 | |
5GP7
 
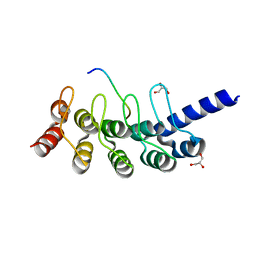 | | Structural basis for the binding between Tankyrase-1 and USP25 | | Descriptor: | GLYCEROL, Tankyrase-1, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Liu, J, Xu, D, Fu, T, Pan, L. | | Deposit date: | 2016-08-01 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | USP25 regulates Wnt signaling by controlling the stability of tankyrases
Genes Dev., 31, 2017
|
|
5CZW
 
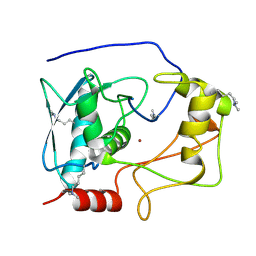 | | Crystal structure of myroilysin | | Descriptor: | Myroilysin, ZINC ION | | Authors: | Zhou, J, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2015-08-01 | | Release date: | 2016-08-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Myroilysin is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Activated by a Cysteine-switch Mechanism
J. Biol. Chem., 292, 2017
|
|
5Y0E
 
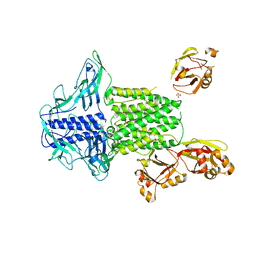 | |
8SLR
 
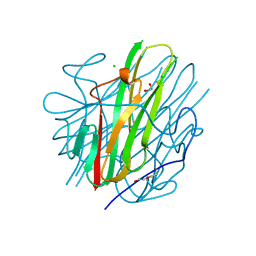 | | Crystal Structure of mouse TRAIL | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pedersen, L.C, Xu, D. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Heparan sulfate promotes TRAIL-induced tumor cell apoptosis.
Elife, 12, 2024
|
|
6P0K
 
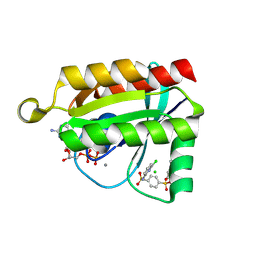 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(6-chloropyridin-2-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P0O
 
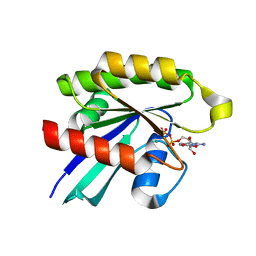 | | Crystal structure of GDP-bound human RalA | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Ral-A | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P0N
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(6-fluoropyridin-2-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P0M
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(3-chloropyridin-2-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P0I
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5-methoxypyridin-3-yl)sulfamoyl]benzene-1-sulfonic acid, ACETATE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P0L
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(2-methoxypyridin-3-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2HG7
 
 | | Solution NMR structure of Phage-like element PBSX protein xkdW, Northeast Structural Genomics Consortium Target SR355 | | Descriptor: | Phage-like element PBSX protein xkdW | | Authors: | Liu, G, Parish, D, Xu, D, Atreya, H, Sukumaran, D, Ho, C.K, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Phage-like element PBSX protein xkdW, Northeast Structural Genomics Consortium Target SR355
TO BE PUBLISHED
|
|
2HFI
 
 | | Solution NMR Structure of Protein yppE from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR213 | | Descriptor: | Hypothetical protein yppE | | Authors: | Liu, G, Singarapu, K.K, Parish, D, Eletsky, A, Xu, D, Sukumaran, D, Ho, C.K, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-23 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of hypothetical protein yppE: Northeast Structural Genomics Consortium Target SR213
TO BE PUBLISHED
|
|
8V57
 
 | | Complex of murine cathepsin K with bound cystatin C inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin K, ... | | Authors: | Pedersen, L.C, Xu, D. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Heparan sulfate selectively inhibits the collagenase activity of cathepsin K.
Matrix Biol., 129, 2024
|
|
8V58
 
 | | Complex of murine cathepsin K with bound heparan sulfate 12mer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Pedersen, L.C, Xu, D, Krahn, J.M. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Heparan sulfate selectively inhibits the collagenase activity of cathepsin K.
Matrix Biol., 129, 2024
|
|
2JPU
 
 | | solution structure of NESG target SsR10, Orf c02003 protein | | Descriptor: | Orf c02003 protein | | Authors: | Wu, Y, Singarapu, K.K, Zhang, Q, Eletski, A, Xu, D, Sukumaran, D, Parish, D, Wang, D, Jiang, M, Cunningham, K, Maglaqui, M, Owens, L, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-23 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of NESG target SsR10, Orf c02003 protein
To be Published
|
|
5XNB
 
 | |
3S1S
 
 | | Characterization and crystal structure of the type IIG restriction endonuclease BpuSI | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Shen, B.W, Xu, D, Chan, S.-H, Zheng, Y, Zhu, Y, Xu, S.-Y, Stoddard, B.L. | | Deposit date: | 2011-05-16 | | Release date: | 2011-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI.
Nucleic Acids Res., 39, 2011
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
2JVM
 
 | | Solution NMR structure of Rhodobacter sphaeroides protein RHOS4_26430. Northeast Structural Genomics Consortium target RhR95 | | Descriptor: | Uncharacterized protein | | Authors: | Eletsky, A, Sukumaran, D, Zhang, Q, Parish, D, Xu, D, Wang, H, Janjua, H, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-21 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Rhodobacter sphaeroides protein RHOS4_26430.
To be Published
|
|
3F5F
 
 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | Authors: | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5CIO
 
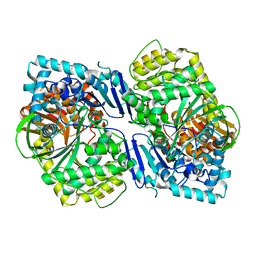 | | Crystal structure of PqqF | | Descriptor: | ZINC ION, pyrroloquinoline quinone biosynthesis protein PqqF | | Authors: | Wei, Q, Xu, D, Ran, T, Wang, W. | | Deposit date: | 2015-07-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
J.Biol.Chem., 291, 2016
|
|
6LR0
 
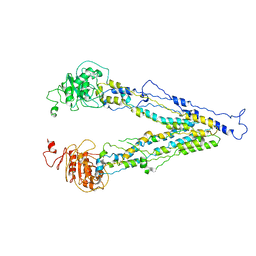 | | structure of human bile salt exporter ABCB11 | | Descriptor: | Bile salt export pump | | Authors: | Wang, L, Hou, W.T, Chen, L, Jiang, Y.L, Xu, D, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human bile salts exporter ABCB11.
Cell Res., 30, 2020
|
|
