6H4M
 
 | | TarP-UDP-GlcNAc-3RboP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable ss-1,3-N-acetylglucosaminyltransferase, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-07-22 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
6H4F
 
 | | TarP-3RboP | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
6H21
 
 | | TarP-UDP-GlcNAc-Mn | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
6HNQ
 
 | | TarP-6RboP-(CH2)6NH2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable ss-1,3-N-acetylglucosaminyltransferase, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-09-17 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
2HIW
 
 | |
4DZO
 
 | |
3PNG
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((2-fluoro-2-(3-fluorophenyl)ethyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(2R)-2-fluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S, Poulos, T.L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Improved Synthesis of Chiral Pyrrolidine Inhibitors and Their Binding Properties to Neuronal Nitric Oxide Synthase.
J.Med.Chem., 54, 2011
|
|
3PNH
 
 | | Structure of Bovine Endothelial Nitric Oxide Synthase Heme Domain in complex with 6-(((3R,4R)-4-(2-((2-FLUORO-2-(3-FLUOROPHENYL) ETHYL)AMINO)ETHOXY)PYRROLIDIN-3-YL)METHYL)-4-METHYLPYRIDIN-2-AMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(2R)-2-fluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Improved Synthesis of Chiral Pyrrolidine Inhibitors and Their Binding Properties to Neuronal Nitric Oxide Synthase.
J.Med.Chem., 54, 2011
|
|
3PNF
 
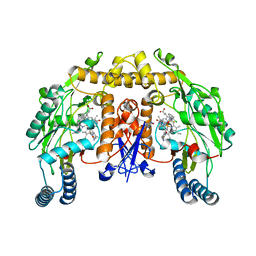 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((2,2-Difluoro-2-(2-chlorophenyl)ethyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2-(2-chlorophenyl)-2,2-difluoroethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S, Poulos, T.L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Improved synthesis of chiral pyrrolidine inhibitors and their binding properties to neuronal nitric oxide synthase.
J.Med.Chem., 54, 2011
|
|
3PNE
 
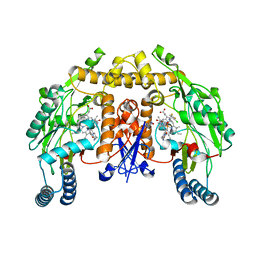 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((2,2-Difluoro-2-(3-chlorophenyl)ethyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2-(3-chlorophenyl)-2,2-difluoroethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S, Poulos, T.L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Improved synthesis of chiral pyrrolidine inhibitors and their binding properties to neuronal nitric oxide synthase.
J.Med.Chem., 54, 2011
|
|
7XGO
 
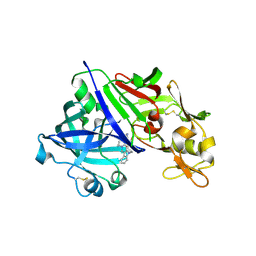 | | Human renin in complex with compound2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
7XGK
 
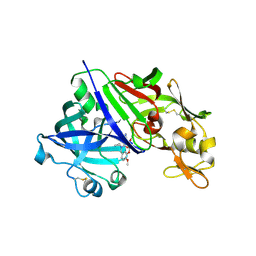 | | Human renin in complex with compound1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
7XGP
 
 | | Human renin in complex with compound3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of SPH3127: A Novel, Highly Potent, and Orally Active Direct Renin Inhibitor.
J.Med.Chem., 65, 2022
|
|
3R21
 
 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7A8W
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with an engineered HMA domain of Pikp-1 from rice (Oryza sativa) | | Descriptor: | NBS-LRR class disease resistance protein, Uncharacterized protein | | Authors: | Maidment, J.H.R, De la Concepcion, J.C, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
8IFQ
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFS
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 7 | | Descriptor: | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGX
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 9 (simnotrelvir, SIM0417, SSD8432) | | Descriptor: | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8THL
 
 | |
8THK
 
 | |
6TYM
 
 | | KEAP1 Kelch domain in complex with Compound 9 | | Descriptor: | (3S)-3-[2-(benzenecarbonyl)-5-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl]-3-(1-ethyl-4-methyl-1H-benzotriazol-5-yl)propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.422 Å) | | Cite: | Design, synthesis and identification of novel, orally bioavailable non-covalent Nrf2 activators.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
