2YJY
 
 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
9BVT
 
 | | RNA Pol II - High Mn(+2) concentration | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Calero, G. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BW0
 
 | | RNA Polymerase II - No ATP | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Calero, G. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
2PVC
 
 | | DNMT3L recognizes unmethylated histone H3 lysine 4 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, Histone H3 peptide, ZINC ION | | Authors: | Cheng, X. | | Deposit date: | 2007-05-09 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Nature, 448, 2007
|
|
7CU9
 
 | | Crystal structure of the soluble domain of TiME protein from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tube-forming protein in Mycobacterial Envelpe, ... | | Authors: | Gong, W, Cai, X, Liu, L, Wen, C. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and architecture of a putative secretion tube across mycobacterial outer envelope.
Sci Adv, 7, 2021
|
|
7CU8
 
 | | Crystal structure of the soluble domain of TiME protein from Mycobacterium tuberculosis | | Descriptor: | SULFATE ION, Tube-forming protein in Mycobacterial Envelope (TiME) | | Authors: | Gong, W, Cai, X, Liu, L, Wen, C. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification and architecture of a putative secretion tube across mycobacterial outer envelope.
Sci Adv, 7, 2021
|
|
6NHW
 
 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
2PV0
 
 | | DNA methyltransferase 3 like protein (DNMT3L) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, ZINC ION | | Authors: | Cheng, X. | | Deposit date: | 2007-05-09 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Nature, 448, 2007
|
|
8U9R
 
 | | STRUCTURAL BASIS OF TRANSCRIPTION: RNA POLYMERASE II SUBSTRATE BINDING AND METAL COORDINATION USING A FREE-ELECTRON LASER | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Arjunan, P, Calero, G, Kaplan, C.D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U9X
 
 | | STRUCTURAL BASIS OF TRANSCRIPTION: RNA POLYMERASE II SUBSTRATE BINDING AND METAL COORDINATION AT 3.0 A OF T834P MUTANT USING A FREE-ELECTRON LASER | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Arjunan, P, Calero, G, Kaplan, C.D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XGS
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8XGU
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8XGO
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
6DUN
 
 | | Crystal Structure Analysis of PIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, TRIHYDROXYARSENITE(III) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells.
Nat Commun, 9, 2018
|
|
8HMV
 
 | | Structure of GPR21-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Wong, T.S, Gao, W. | | Deposit date: | 2022-12-05 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of orphan G protein-coupled receptor GPR21.
MedComm (2020), 4, 2023
|
|
8IJA
 
 | | Cryo-EM structure of human HCAR2-Gi complex with niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-26 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJD
 
 | | Cryo-EM structure of human HCAR2-Gi complex with MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJ3
 
 | | Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJB
 
 | | Cryo-EM structure of human HCAR2-Gi complex with acipimox | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8ZR5
 
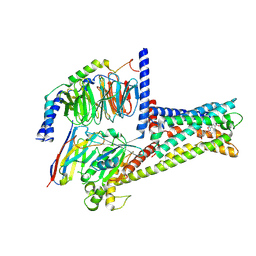 | | Cryo-EM Structure of GPR119-Gs-Firuglipel complex | | Descriptor: | 4-[5-[(1~{R})-1-(4-cyclopropylcarbonylphenoxy)propyl]-1,2,4-oxadiazol-3-yl]-2-fluoranyl-~{N}-[(2~{R})-1-oxidanylpropan-2-yl]benzamide, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To Be Published
|
|
8ZRK
 
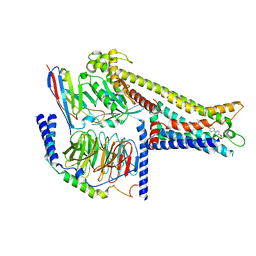 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist GSK-1292263 | | Descriptor: | 5-[4-[[6-(4-methylsulfonylphenyl)pyridin-3-yl]oxymethyl]piperidin-1-yl]-3-propan-2-yl-1,2,4-oxadiazole, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Xiong, T.T, Zeng, Z.C, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To be published
|
|
3OAP
 
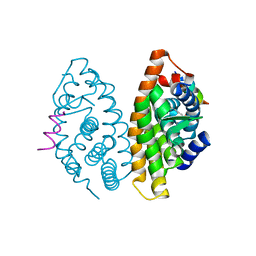 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 9-cis retinoic acid and the coactivator peptide GRIP-1 | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, Energetics and Dynamics of Binding Coactivator Peptide to Human Retinoid X Receptor Alpha Ligand Binding Domain Complex with 9-cis-Retinoic Acid.
Biochemistry, 50, 2011
|
|
